Chlamydia’s stealthy cloaking device identified
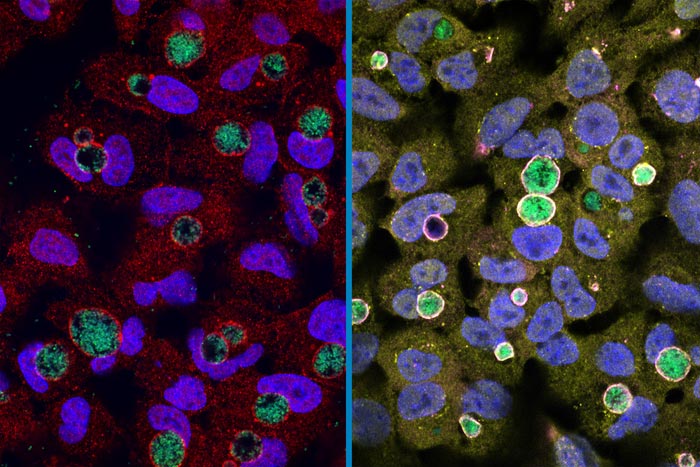
At left: A wild type Chlamydia (green) surrounded by the GarD protein (red) that cloaks it from detection inside human cells. Right: Chlamydia with GarD knocked out (green) are enveloped by the antimicrobial proteins ubiquitin (yellow) and RNF213 (magenta).
Credit: Stephen C. Walsh, Duke University
Microbial proteins around a sexually transmitted infection allow pathogen to hide undetected inside host cells.
Chlamydia, the leading cause of sexually transmitted bacterial infections, evades detection and elimination inside human cells by use of a cloaking device. But Duke University researchers have grasped the hem of that invisibility cloak and now hope they can pull it apart.
To enter the cell and peacefully reproduce, many pathogenic bacteria, including Chlamydia, cloak themselves in a piece of the cell’s membrane, forming an intracellular free-floating bubble called a vacuole or, in the case of Chlamydia, an inclusion. Chlaymydia’s cloak appears to be especially effective at evading the cell’s built-in immunity, allowing the infection to last for months.
A Duke team led by graduate student Stephen Walsh and Jörn Coers, PhD, an associate professor of molecular genetics and microbiology in the Duke School of Medicine, wanted to know how the cloaking worked.
“We knew there was the potential to kill Chlamydia, but when we did experiments with the human-adapted form, Chlamydia trachomatis, it was very good at growing in human cell cultures,” Coers said. Even after the scientists used an immune stimulant to alert the cell’s defense systems of the presence of Chlamydia, nothing happened. “We said, there’s the pathogen. Our defense system should see it. Why does it not see it?”
They ran their experiments again using a mouse-adapted version of the Chlamydia bacteria in human cells to see how the cell’s immune system responded to a non-human pathogen.
“Humans, don’t get mouse Chlamydia because it evolved with mice and human Chlamydia evolved with humans,” Coers said. “So there’s this really fine-tuned adaptation that the pathogen has undergone.” The mouse version of the bacterial inclusion was readily identified and labeled for destruction in human cells.
“Chlamydia trachomatis is so good at evading our human responses,” Coers said. “It still causes an inflammatory disease, but it’s a very slow disease.”
This evolutionary arms race between the immune system and the pathogen has been going on for millions of years. “Mouse and human adapted Chlamydia have a common ancestor,” Coers said. “However, this common ancestor may go back as far as when humans and rodents basically split from each other. This is a long time for the bacteria to really fine-tune their interactions with their host species.”
Working with Duke MGM colleagues Raphael Valdivia and Robert Bastidas, the researchers ran a large genetic screen of Chlamydia that identified a protein, GarD (gamma resistance determinant), that appeared to be blocking the host cell’s ability to mark a Chlamydia inclusion for destruction by the immune system.
Mutating their GarD genes left the bacteria vulnerable. “GarD is the stealth factor,” Coers said.
Specifically, GarD interferes with the ability of a giant signaling protein called RNF213 or mysterin to sense small bits of bacterial molecules poking out of the shell of the inclusion. “RNF213 is basically the eyes of the immune system,” Coers said. Having blinded mysterin in this fashion, the signal for immune flagging and destruction is never started.
The inside of a cell is swarming with these little bubbles of membrane-covered vacuoles; most are friends, but some, like the Chlamydia inclusion, are foes.
“There’s so many different types of membranes and vacuoles that live inside a cell,” Coers said. “How is the immune system able to find the rare vacuole that contains a pathogen? In the case of Chlamydia, we really don’t have the answer to that question. But whatever it is, we believe this enzyme (mysterin) is seeing it.”
Unfortunately, that’s all the further this story goes for now, Coers said. This is a great new insight into a pernicious infection, but several steps away from a therapy. Researchers still need to figure out how mysterin sees those bacterial molecules in the first place and how GarD blinds mysterin.
“If you could find a mechanism to deactivate GarD, then you can turn human Chlamydia into mouse Chlamydia,” Coers said. “That would allow us to harness the powers of our own immune system to clear infection.”
New Chlamydia infections occur in 200,000 Americans per year and are often asymptomatic for months or even years while being transmissible through sexual contact. With time, an untreated infection can lead to pelvic inflammatory disease, ectopic pregnancy and female infertility.
The US Centers for Disease Control recommends that young women be tested for Chlamydia annually.
This research was supported by grants from the National Institutes of Health (A030801, AI150106, AI140019,U19-AI084044) and the Burroughs Wellcome Fund.
CITATION: “The Bacterial Effector GarD Shields Chlamydia Trachomatis Inclusoins from RNF213-Mediated Ubiquitylation and Destruction,” Stephen Walsh, Jeffrey Reitano, Mary Dikinson, Miriam Kutsch, Dulcemaria Hernandez, Alyson Barnes, Benjamin Schott, Liuyan Wang, Dennis Ko, So Young Kim, Raphael Valdivia, Robert Bastidas, Jörn Coers. Cell Host & Microbe, Nov. 9, 2022.
Journal: Cell Host & Microbe
Method of Research: Experimental study
Subject of Research: Animals
Article Title: The Bacterial Effector GarD Shields Chlamydia Trachomatis Inclusoins from RNF213-Mediated Ubiquitylation and Destruction
Article Publication Date: 9-Nov-2022
Media Contact
Karl Bates
Duke University
karl.bates@duke.edu
Office: 919-681-8054
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Properties of new materials for microchips
… can now be measured well. Reseachers of Delft University of Technology demonstrated measuring performance properties of ultrathin silicon membranes. Making ever smaller and more powerful chips requires new ultrathin…

Floating solar’s potential
… to support sustainable development by addressing climate, water, and energy goals holistically. A new study published this week in Nature Energy raises the potential for floating solar photovoltaics (FPV)…

Skyrmions move at record speeds
… a step towards the computing of the future. An international research team led by scientists from the CNRS1 has discovered that the magnetic nanobubbles2 known as skyrmions can be…





















