When one reference genome is not enough

A single reference genome is not enough to harness the full genetic variation of a species so pan-genomes of crops would be extremely useful. The phenotypic diversity of Brachypodium plants is demonstrated in this image, which is associated with a news release for a Nature Communications paper in which an international team led by DOE Joint Genome Institute researchers gauged the size of a plant pan-genome using the model grass Brachypodium distachyon. Credit: John Vogel
Having plant pan-genomes for crops that are important for fuel and food applications would enable breeders to harness natural diversity to improve traits such as yield, disease resistance, and tolerance of marginal growing conditions.
In a paper published December 19, 2017 in Nature Communications, an international team led by researchers at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility at Lawrence Berkeley National Laboratory (Berkeley Lab), gauged the size of a plant pan-genome using Brachypodium distachyon, a wild grass widely used as a model for grain and biomass crops. As one of the JGI's Plant Flagship Genomes, B. distachyon ranks among the most complete plant reference genomes.
“There are a vast number of genes that are not captured in a single reference genome,” added study senior author John Vogel, head of the JGI's Plant Functional Genomics group. “Indeed, about half of the genes in the pan-genome are found in a variable number of lines.” Working toward the primary goal of accurately estimating the size of a plant pan-genome, Vogel and his colleagues performed whole-genome de novo assembly and annotation of 54 geographically diverse lines of B. distachyon, yielding a pan-genome containing nearly twice the number of genes found in any individual line.
“The genome of a species is a collection of genomes, each with their own unique twist,” added JGI bioinformaticist and study first author Sean Gordon. “Now knowing that focusing on a single reference genome leads to incomplete and biased estimates of genetic diversity and ignores genes potentially important for breeding applications, we should better incorporate multiple references in future studies of natural diversity.”
Moreover, genes found in only some lines tend to contribute to biological processes (e.g., disease resistance, development) that may be beneficial under some environmental conditions, whereas genes found in every line usually underpin essential cellular processes (e.g., glycolysis, iron transport).
“This means that the variable genes are being preferentially retained if they are beneficial under some conditions. These are exactly the types of genes that breeders need to improve crops.” Vogel said.
In addition, genes found in only a subset of lines displayed faster rates of evolution, lay closer to transposable elements (thought to play a key role in pan-genome evolution), and were less likely to be found in the same chromosomal location as functionally equivalent genes in other grasses.
The sequence assemblies, gene annotations and related information can be downloaded from the project website BrachyPan: brachypan.jgi.doe.gov. The Brachypodium distachyon genome is available on the JGI Plant Portal Phytozome: phytozome.jgi.doe.gov.
###
Key collaborators on the project include Pilar Catalan and Bruno Contreras-Moreira with the University of Zaragoza in Spain, who performed population and evolutionary analysis, and clustering genes to help create the pan-genome, respectively. Additional key collaborators include Richard Amasino and Daniel Woods with the University of Wisconsin-Madison and the Great Lakes Bioenergy Research Center, a DOE Bioenergy Research Center, who correlated pan-genes with flowering and vernalization phenotypes.
Other JGI personnel who contributed to the study include: Shengqiang Shu, who annotated the genomes and looked for missed pan-genes in the reference; Wendy Schackwitz, Joel Martin, and Anna Lipzen, who detected variants with respect to the reference genome; Jeremy Phillips, who helped create gene families; Kerrie Berry, who managed the genome sequencing project; and David Goodstein and Patrick Davidson, who designed and created the BrachyPan website.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
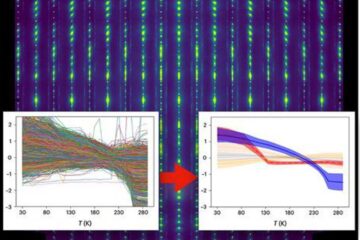
Machine learning algorithm reveals long-theorized glass phase in crystal
Scientists have found evidence of an elusive, glassy phase of matter that emerges when a crystal’s perfect internal pattern is disrupted. X-ray technology and machine learning converge to shed light…
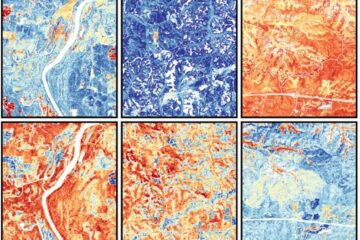
Mapping plant functional diversity from space
HKU ecologists revolutionize ecosystem monitoring with novel field-satellite integration. An international team of researchers, led by Professor Jin WU from the School of Biological Sciences at The University of Hong…

Inverters with constant full load capability
…enable an increase in the performance of electric drives. Overheating components significantly limit the performance of drivetrains in electric vehicles. Inverters in particular are subject to a high thermal load,…





















