'Resurrection ecology' of 600-year-old water fleas used to understand pollution adaptation
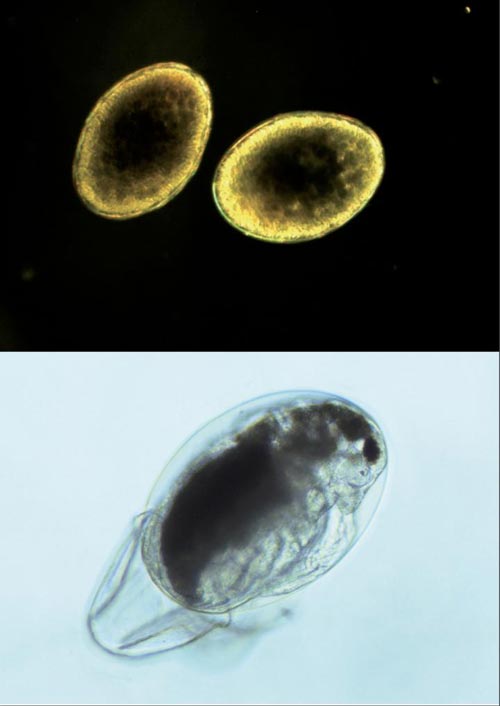
In this picture, a hatching Daphnia (bottom) and a pair of dormant Daphnia eggs (top-isolated from an ephippium, a kind of protective case produced by the 'mother Daphnia' in which the eggs get buried in the sediment. Credit: Dagmar Frisch
One of the leading threats to lakes since the rise of agriculture are runoffs from fertilizer, in the form of high phosphorus levels. These can trigger devastating events like eutrophication, where deadly algal blooms thrive on phosphorus, and in the process, outcompete and choke off vital nutrients from the rest of the lake.
But to best learn how organisms can adapt to eutrophication events requires comparing samples before and after the event —a seemingly impossible feat to study eutrophication—since it arose with modern agricultural 100 years ago.
So, by taking advantage of the unique genomic model organism of tiny waterfleas, or Daphnia, an international team of researchers has now analyzed Daphnia from a phosphorus-rich Minnesota lake —and compared it to revived, 600-year-old Daphnia dormant eggs found in the bottom sediments—- to better understand how these creatures cope with a dramatic environmental change.
Dagmar Frisch and her colleagues, based in the University of Birmingham's School of Biosciences, used the 'resurrection ecology' of Daphnia and new analysis tools to perform the study. The team was only able to make these discoveries by comparing the responses of modern Daphnia with their 600-year-old ancestors.
Both the modern and the ancient samples studied came from the same lake in Minnesota where eutrophication first started at the beginning of the 20th century. “We used existing data and state-of-the-art analytical methods to connect patterns of gene expression with the physiological responses that allow these animals to deal with increased environmental phosphorus” said author Dagmar Frisch, an expert in environmental paleogenomics. “This allowed us to identify which part of the gene network was accountable for the newly evolved response.”
“Because Daphnia is such a central species in aquatic ecosystems, our study ultimately improves our understanding of how aquatic ecosystems can mitigate some of the effects of eutrophication, one of the major global threats to freshwater environments,” said co-author Dörthe Becker, an expert in environmental 'omics'.
They were able to show a large cluster of several hundred genes uniquely adapted in modern-day Daphnia to high phosphorus levels. Many of these were involved in vital, core metabolic pathways necessary for Daphnia survival.
“We used network analysis methods to find out which genes 'communicate' with others to form clusters (or “modules”), and how this gene communication has changed in a keystone species over the last 600 years. In addition, we were able to connect these modules with particular observed traits, which was achieved for the first time in resurrection ecology,” said co-author Marcin Wojewodzic.
“Our study emphasizes that evolution is a result of molecular fine-tuning that happens on different layers, ranging from basic cellular responses to complex physiological traits” said Becker. “The approach we used allows a more holistic view of how animals can and do respond to environmental change, and by that improve our understanding of organisms as integrated units of biological organization,” said Frisch.
Next, the team will continue to explore how these networks and other molecular processes, including epigenetics play a role in evolutionary adaptation to changing environments.
###
The study was published recently in the advanced online edition of Molecular Biology and Evolution.
Adapted from a story by Beck Lockwood, University of Birmingham r.lockwood@bham.ac.uk
Media Contact
More Information:
http://dx.doi.org/10.1093/molbev/msz234All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
Solving the riddle of the sphingolipids in coronary artery disease
Weill Cornell Medicine investigators have uncovered a way to unleash in blood vessels the protective effects of a type of fat-related molecule known as a sphingolipid, suggesting a promising new…

Rocks with the oldest evidence yet of Earth’s magnetic field
The 3.7 billion-year-old rocks may extend the magnetic field’s age by 200 million years. Geologists at MIT and Oxford University have uncovered ancient rocks in Greenland that bear the oldest…

Decisive breakthrough for battery production
Storing and utilising energy with innovative sulphur-based cathodes. HU research team develops foundations for sustainable battery technology Electric vehicles and portable electronic devices such as laptops and mobile phones are…





















