New Data Suggests Current Approach to Drug Discovery for Lou Gehrig’s Disease Be Re-Examined

> Researchers at the University of Pennsylvania School of Medicine have found an absolute biochemical distinction between the sporadic and hereditary variants of Lou Gehrig’s disease, or amyothrophic lateral sclerosis (ALS), suggesting that current approaches to drug discovery should be re-examined.
> By examining the various forms of ALS in post-mortem tissue, the researchers found that TDP-43 was the disease protein in sporadic ALS cases, but not in patients with SOD-1 mutations, all of whom have the familial form of ALS. Patients with the SOD-1 mutation account for about 1 percent of all ALS cases.
> This finding may partially account for why therapeutic strategies, shown to be effective in SOD-1 mouse models, have generally not been effective in clinical trials of sporadic ALS patients.
> The study was published this week in the Annals of Neurology.
Most research on Lou Gehrig’s disease therapeutics has been based on the assumption that its two forms (sporadic and hereditary) are similar in their underlying cause. Now, researchers at the University of Pennsylvania School of Medicine have found an absolute biochemical distinction between these two disease variants, suggesting that current approaches to drug discovery should be re-examined.
About 5 percent of all cases of Lou Gehrig’s disease, or amyothrophic lateral sclerosis (ALS), are passed from generation to generation. The most common genetic variant in this familial form is caused by a mutation in the SOD-1 gene. The researchers looked at a large set of ALS patients, including hereditary cases, both with and without the SOD-1 mutation.
The present study – published this week in the Annals of Neurology – was conducted by Penn; a group led by Ian Mackenzie from the University of British Columbia; the University of Munich; and others across the U.S. and Canada.
“Most ALS research has focused on how mutant SOD-1 proteins are toxic to nerve cells,” says senior author John Trojanowski, MD, PhD, who directs the Penn Institute on Aging. Last year, Penn investigators led by co-author Virginia Lee, PhD, who directs the Penn Center for Neurodegenerative Disease Research, identified TDP-43 as the major disease protein in sporadic (non-hereditary) forms of ALS, which are not those caused by SOD-1 gene mutations.
By examining the various forms of ALS in post-mortem tissue, the researchers found that TDP-43 was the disease protein in sporadic ALS cases, but not in patients with SOD-1 mutations, all of whom have the familial form of ALS. Patients with the SOD-1 mutation account for about 1 percent of all ALS cases.
“We argue that SOD-1 ALS does not equal sporadic ALS,” says Trojanowski. “If you pursue drug discovery focusing on SOD-1-mediated pathways of brain and spinal cord degeneration you may benefit SOD-1-bearing patients, but not the vast majority of ALS patients who have the sporadic form of this disorder with TDP-43 pathologies underlying the disease.”
“Motor neuron degeneration in TDP-43 cases may result from a different mechanism than cases with SOD-1 mutations, so this form of ALS may not be the familial counterpart of sporadic ALS,” surmises Lee.
“This may also partially account for why therapeutic strategies, shown to be effective in SOD-1 mouse models, have generally not been effective in clinical trials of sporadic ALS patients,” explains Trojanowski. “This also sounds a cautionary note in all other diseases in which you have familial and sporadic versions of the disease because it will prompt researchers to ask if mouse models for drug discovery are based on the correct mutations or disease protein.”
This research was funded by the Canadian Institutes of Health Research, the National Institute on Aging, the German Federal Ministry of Education and Research, The Wellcome Trust (United Kingdom) and the UK Medical Research Council.
Co-authors in addition to Trojanowski, Lee, and Mackenzie are Eileen H. Bigio, Paul G. Ince, Felix Geser, Manuela Neumann, Nigel J. Cairns, Linda K. Kwong, Mark S. Forman, John Ravits, Heather Stewart, Andrew Eisen, Leo McClusky, Hans A. Kretzschmar, Camelia M. Monoranu, J. Robin Highley, Janine Kirby, Teepu Siddique, and Pamela J. Shaw.
Media Contact
More Information:
http://www.uphs.upenn.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
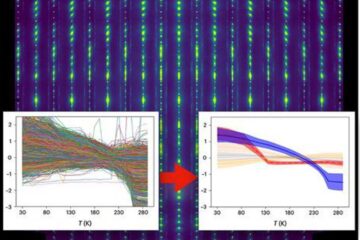
Machine learning algorithm reveals long-theorized glass phase in crystal
Scientists have found evidence of an elusive, glassy phase of matter that emerges when a crystal’s perfect internal pattern is disrupted. X-ray technology and machine learning converge to shed light…
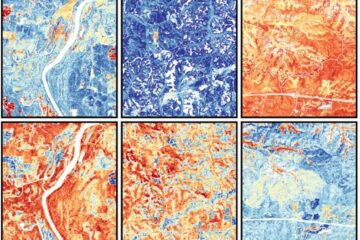
Mapping plant functional diversity from space
HKU ecologists revolutionize ecosystem monitoring with novel field-satellite integration. An international team of researchers, led by Professor Jin WU from the School of Biological Sciences at The University of Hong…

Inverters with constant full load capability
…enable an increase in the performance of electric drives. Overheating components significantly limit the performance of drivetrains in electric vehicles. Inverters in particular are subject to a high thermal load,…





















