North and South American Researchers Find Architectural Abnormalities in T. Cruzi Genome
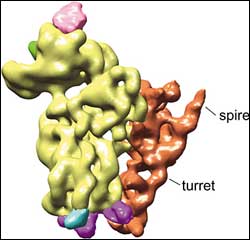
T. cruzi ribosome <br>Unusual turret and spire features of the small subunit of the T. cruzi ribosome. <br>Image: Joachim Frank Lab, Adapted from PNAS
A Howard Hughes Medical Institute investigator from Albany, New York, and an HHMI international research scholar from Buenos Aires, Argentina have combined their expertise to identify two peculiar features of the protein-making machinery of the parasite that causes Chagas disease. Their findings could help scientists develop a safe and effective drug for the disease, whose cardiac complications kill up to 30 percent of those infected.
The unusual structure of the ribosome, published online the week of July 4, 2005, in the Proceedings of the National Academy of Sciences, also suggests that the parasite has a unique scanning mechanism for translating genetic information into proteins.
“This may lead the way to the discovery or development of drugs that are specific against an essential mechanism of the life of the parasite and do not affect the infected organism,” said parasitologist Mariano Levin, an HHMI international research scholar at the Institute for Research on Genetic Engineering and Molecular Biology in Buenos Aires and co-author of the study.
Levin met senior author Joachim Frank at a science meeting at HHMI headquarters in Chevy Chase, Maryland. Frank is an HHMI investigator at Health Research Inc., at the Wadsworth Center in Albany, New York, who had pioneered the analysis of ribosome structures by a method he developed for three-dimensional reconstruction of large molecules using electron microscopy and image processing. Levin told him the story of Chagas and the need to know more about the structure of the parasite’s ribosome.
A ribosome is basically a double ball made mostly of RNA and over 50 proteins. The balls, which are of unequal size, are called small and large subunits. After a string of messenger RNA (mRNA) copies the information encoded on a gene, it enters a sea of ribosomes, where that genetic information is translated into a protein product.
Scientists have long known there was something strange about the genetic translational machinery of the Chagas parasite. Unlike in other organisms, an identical string of 39 nucleotides is spliced onto the lead end of every mRNA molecule, no matter which protein is to be made. And, as Levin and his colleagues discovered years ago, the heart damage associated with Chagas disease is triggered by antibodies formed by an infected individual’s immune system in response to a unique ribosomal protein from the parasite.
Chagas disease exists only on the American continent, according to the World Health Organization. It is caused by a parasite called Trypanosoma cruzi, which is transmitted to humans by a blood-sucking bug usually found in poor areas with substandard housing. An estimated 16 to 18 million people in Latin America are infected, and another 100 million are at risk. Some cases have spread through blood transfusion. Screening blood for the disease is mandatory in many Latin American countries, where more blood is infected by the parasite than by HIV and hepatitis B and C, but not in the United States, where the disease is rare.
For a few people, symptoms may begin within a few days or weeks. Most people may not know they have the disease for a decade or more, during which time 20 to 30 percent will develop potentially deadly cardiac problems. According to Levin, there is an urgent need for a more reliable treatment for this chronic phase of the infection.
In addition to the public health implications, Frank sees the parasite as a primitive link to modern eukaryotes and its ribosome as a source of potential insights into the evolution of more complex organisms.
“Ribosomes perform the translation of genetic information into protein in all organisms,” Frank said. “The principle is always the same in plants, bacteria, and humans. It’s a fundamental property of life on Earth.”
Another HHMI investigator, Thomas A. Steitz of Yale University, produced the first atomic-resolution images of the large subunit of the ribosome from a bacterium. He reported on the work in a landmark paper in 2000 in the journal Science. Other groups solved the structure of the small subunit as well.
To date, however, no atomic-resolution image of eukaryotic ribosomes exists, and all images produced so far have been the result of Frank’s electron microscope imaging and reconstruction methods.
The ribosomes of different eukaryotic organisms have only slight differences in their peripheral elements, such as specialized tools to send newly formed proteins into or through membranes. If anything, Frank expected the trypanosome’s ribosome might be missing some elements, because of its sheltered life within host organisms.
“In reconstructions of ribosomes from yeast to rabbits to humans, one essentially sees the same architecture,” said Frank. “It lulled us into the belief that once you’ve seen one ribosome, you’ve seen them all. It was a total surprise to see such a different structure.”
Last year, Levin traveled to Frank’s lab in New York with purified ribosomes from T. cruzi. There, postdoctoral fellow Haixiao Gao led the experiment, using a technique that works on a principle similar to a medical CAT scan.
First, the researchers freeze an ultrathin layer of buffer with purified ribosomes so quickly that the water has no chance to crystallize and damage the ribosomes. Then, the frozen sample is imaged with an electron beam in the transmission electron microscope.
A CAT scan takes readings of a single body from different angles to reconstruct its three-dimensional images, but the microscope catches hundreds of ribosomes at different angles in a single snapshot. Frank and his colleagues have developed a battery of complex mathematical techniques to reconcile the images at different angles. From a total of up to 30,000 ribosome images, they assemble a final three-dimensional image of a single ribosome.
A ribosome binds to mRNA with its small subunit. In the new study, the most peculiar feature showed up on the small subunit of the parasite’s ribosome.
The researchers found a gigantic addition tacked onto one side of the small subunit, resembling a turret topped by a spire. “It’s larger than anything we’ve seen anywhere, as far as peripheral additions in ribosomes are concerned,” Frank said. “It’s almost freestanding.”
The exact purpose of the turret and its spire is unknown, but the researchers suspect the function will be as unique as its architecture. They speculate that it may have something to do with the 39-nucleotide leader of the mRNA that the parasite’s ribosomes must process. The spot where the ribosome binds to the mRNA, for example, is roughly as far away from the turret and spire as the length of the 39-nucleotide leader sequence.
“It could provide a unique scanning mechanism different from other eukaryotes,” Frank said. “For instance, the splice leader of the mRNA might attach at the turret, which may swivel out and drag the mRNA across the binding site on the ribosome to initiate protein translation.”
The findings may also advance studies of parasitic relatives Trypanosoma brucei, which causes sleeping sickness in Africa, and Leishmania, a parasite spread by sand flies. Higher resolution models of the ribosome structure will be needed to design targeted drugs, Frank said.
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Properties of new materials for microchips
… can now be measured well. Reseachers of Delft University of Technology demonstrated measuring performance properties of ultrathin silicon membranes. Making ever smaller and more powerful chips requires new ultrathin…

Floating solar’s potential
… to support sustainable development by addressing climate, water, and energy goals holistically. A new study published this week in Nature Energy raises the potential for floating solar photovoltaics (FPV)…

Skyrmions move at record speeds
… a step towards the computing of the future. An international research team led by scientists from the CNRS1 has discovered that the magnetic nanobubbles2 known as skyrmions can be…





















