A quick-change artist: tiny protein folds faster than any other
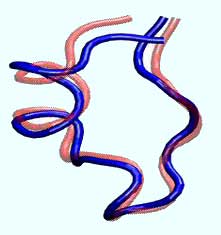
University of Florida researchers have discovered that the Tryptophan cage protein, derived from the saliva of the Gila monster lizard, zooms to its folded state, above, in four millionths of a second - about four times faster than any protein previously measured. The finding adds to the emerging knowledge about how proteins fold, information that could lead to better drugs and cures for diseases tied to misshapen proteins, such as Alzheimer’s, Parkinson’s and Mad Cow diseases.
The world speed record for protein folding apparently goes to an unusually tiny specimen that traces its origins to Gila monster spit.
So reports a team of University of Florida researchers in a paper published this week in the online edition of the Journal of the American Chemical Society. Though significant mainly from a purely scientific standpoint, the finding eventually may be important in researchers’ understanding of the underlying causes behind a host of maladies.
Proteins acquire their three-dimensional, blob-like shapes when the amino acids they are composed of spontaneously fold into place. The process has become a hot topic in science in recent years because the shape of proteins is directly tied to their function in the cells of animals and people. Misshapen proteins, or proteins whose amino acids form an even slightly different configuration than normal proteins, have been connected to Alzheimer’s disease and a range of other serious disorders.
The UF team found the protein Tryptophan cage, or Trp-cage for short, rockets from its two-dimensional, line-like state of 20 amino acids to its three-dimensional state in four-millionths of a second. That’s the fastest rate yet observed for a complete protein – and about four times faster than any other protein yet measured, UF researchers say.
With about 10 atoms per amino acid, the protein is composed of about 200 atoms, and each atom must interact with every other atom before finding its proper place in the structure. That means at least 40,000 atomic interactions – pushing and pulling movements – occur in an almost imperceptible period, said Stephen Hagen, an assistant professor of physics and one of the paper’s four UF authors.
“The fact that some proteins can fold incredibly fast is really a remarkable thing,” he said. “What is it that’s special about these molecules that enables them to solve a very difficult computational problem spontaneously in such a short amount of time?”
Vijay Pande, an assistant professor of chemistry at Stanford University, called the UF finding “really important and very exciting.” He said it could speed up biologists’ efforts to simulate the protein-folding process, which could lead to better drugs and cures for diseases tied to misshapen proteins.
Scientists have long known that instructions in genes’ DNA determine the amino acid code for proteins. However, they still don’t know the structure of most human proteins or the role they play in many inherited traits or diseases. The way amino acids come together to form proteins is one area researchers are plumbing for answers.
Enter the Gila monster. Trp-cage stems from a protein another group of researchers removed from the lizard’s saliva in an effort to understand why its bite makes some people ill but not others, said Adrian Roitberg, a UF assistant professor of chemistry. The researchers modified the protein’s structure to make it more stable and easier to work with, and then published the results of their work online, where the UF scientists learned about them.
With other proteins composed of hundreds or thousands of amino acids, Trp-cage’s small size might seem to explain its fast-folding speed, but protein size and speed are not related, Hagen said. More interestingly, researchers expected Trp-cage would fold at least 1,000 times slower than it does, leaving its blinding speed “quite a mystery,” Hagen said.
There are two ways of probing how proteins attain their shape: experiments in the lab and computer simulations. UF researchers have done both with Trp-cage.
Hagen’s team, which included Roitberg and UF physics doctoral students Linlin Qiu and Suzette Pabit, used an advanced instrument called a laser temperature jump spectrometer to observe and time Trp-cage’s transition from its unfolded to its folded state. Roitberg also was part of a separate team collaborating with researchers from the State University of New York-Stonybrook that simulated Trp-cage’s structure on a computer based solely on its amino acid code. The results, reported last month in the Journal of the American Chemical Society, caused a stir in the scientific community because the simulated Trp-cage was extremely close in size and shape to that of the actual observed protein.
If such a computational method ever could be used to replicate larger, more-complex human proteins, it could speed the pace of research dramatically because the laboratory experimental approach is difficult, time consuming and expensive, Roitberg and Hagen said. For now, however, such a goal is far off, because computers are not yet powerful enough to quickly process all the information about each atom’s forces on all of the other atoms in larger proteins.
Roitberg’s team’s simulation of tiny Trp-cage required 16 computers and three weeks of computing time – another indication of the protein’s speedy folding rate. Although protein fragments have been observed to fold faster, the complete Trp-cage is one of a kind. “Here’s a molecule that is able to do in four microseconds what it takes these computers several weeks to do,” Hagen said.
Hagen said many diseases are tied to misshapen proteins. These include Alzheimer’s, Parkinson’s disease, Mad Cow Disease and others, Pande said. For biomedical researchers interested in genetic therapy to correct these proteins’ shapes, that naturally raises the question of how proteins mis-fold into botched versions. So while the news about Trp-cage’s folding pace has no immediate biomedical application, it contributes to increasing knowledge about this important process, Hagen said.
Media Contact
More Information:
http://www.ufl.edu/All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Properties of new materials for microchips
… can now be measured well. Reseachers of Delft University of Technology demonstrated measuring performance properties of ultrathin silicon membranes. Making ever smaller and more powerful chips requires new ultrathin…

Floating solar’s potential
… to support sustainable development by addressing climate, water, and energy goals holistically. A new study published this week in Nature Energy raises the potential for floating solar photovoltaics (FPV)…

Skyrmions move at record speeds
… a step towards the computing of the future. An international research team led by scientists from the CNRS1 has discovered that the magnetic nanobubbles2 known as skyrmions can be…





















