Oregon team says physics explains protein unpredictability
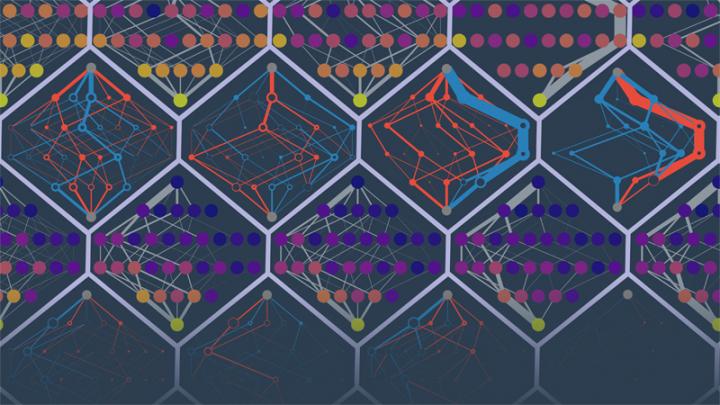
Researchers in the University of Oregon lab of Michael Harms predicted the evolutionary paths of different simulated proteins. The points in the image are protein sequences. The edges denote probabilities of trajectories. Colors denote low to high fitness (purple to yellow) or falsely inaccessible or falsely accessible routes (red and blue). Illustration by Michael Harms and Zach Sailer
University of Oregon scientists theorized that they could manipulate a protein one mutation at a time and predict its evolution. They sought to prove it. And failed.
They do think, however, that they've found a fundamental truth underlying unpredictability in a biological system. Basic physical limitations make uncertainty the norm, they reported in a paper published online Oct. 23 in the Proceedings of the National Academy of Sciences.
“While we got a surprising negative result, we were able to say why,” said Michael J. Harms, a professor in the UO Department of Chemistry and Biochemistry and scientist in the Institute of Molecular Biology. “That is a positive. Our simple study provides confirmation of what many people in the field have observed repeatedly — unpredictability. It appears it is universal.”
The research was a digital affair, done with computer simulations designed by UO doctoral student Zachary R. Sailer. He and Harms created a simple lattice protein, using an approach previously created in the Harms lab, with a random sequence of 12 amino acids. They then ran evolutionary simulations to optimize stability, a physical property of the protein.
The goal was to use the effects of all 228 mutations known to be associated with the starting protein to predict these simulated trajectories: which mutation would occur, when, over time. The ability to project ahead faded fast after the first two mutations. After that, the anticipated trajectories went astray amid a growing number of rerouting probabilities.
“The quality of your information actually decays over time,” Sailer said. “As mutations accumulate, the effects of the mutations that you measured start to change so that you can't predict where you are going.”
In their paper, Sailer and Harms suggest that physics, particularly thermodynamics, is at play. Each mutation alters the protein in a small, but nonlinear way. This means that the effect of each mutation depends on all mutations that occurred before.
“I think that what we showed, fundamentally, is that even if you know a lot about a system, about a protein, you cannot predict how it evolves because of the physics of the system,” Harms said. “There are physical rules that limit evolution and its predictability.”
How proteins evolve is a fundamental question in evolutionary biology, from both a philosophical perspective, to learn more about the machinery of biological systems, and for clues that might lead to improved or better drugs.
“Practically,” Harms said, “our research may help us learn how to prevent the evolution of antibiotic resistance in bacteria.” Almost all bacterial borne infections are developing resistance to antibiotics, creating a leading public health concern around the world.
“Rather than studying the effects of all individual mutations,” Sailer added, “maybe we should study random combinations of many mutations. Such an approach might help us predict the evolution of resistance.”
Work is underway in the Harms lab to test for this possibility on real proteins. “We're building computational tools that let us analyze antibiotic resistance datasets, and we are getting hints that a combinatorial approach does work,” Harms said. “It's more complicated than studying individual mutations, but our work shows the individual approach is unlikely to work.”
###
A fellowship to Harms from the Alfred P. Sloan Foundation funded the research. The UO also provided additional research funds to Sailer.
Sources: Michael J. Harms, assistant professor, Department of Chemistry and Biochemistry and Institute of Molecular Biology, 541-346-9002, harms@uoregon.edu, and Zachary R. Sailer, doctoral student, Department of Chemistry and Biochemistry and Institute of Molecular Biology, 541-346-9003, zsailer@uoregon.edu
Note: The UO is equipped with an on-campus television studio with a point-of-origin Vyvx connection, which provides broadcast-quality video to networks worldwide via fiber optic network. There also is video access to satellite uplink and audio access to an ISDN codec for broadcast-quality radio interviews.
Links:
About Michael Harms: http://molbio.
Harm's Lab: https:/
Institute of Molecular Biology: http://molbio.
Department of Chemistry and Biochemistry: https:/
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Superradiant atoms could push the boundaries of how precisely time can be measured
Superradiant atoms can help us measure time more precisely than ever. In a new study, researchers from the University of Copenhagen present a new method for measuring the time interval,…

Ion thermoelectric conversion devices for near room temperature
The electrode sheet of the thermoelectric device consists of ionic hydrogel, which is sandwiched between the electrodes to form, and the Prussian blue on the electrode undergoes a redox reaction…

Zap Energy achieves 37-million-degree temperatures in a compact device
New publication reports record electron temperatures for a small-scale, sheared-flow-stabilized Z-pinch fusion device. In the nine decades since humans first produced fusion reactions, only a few fusion technologies have demonstrated…





















