How cells dispose of their waste
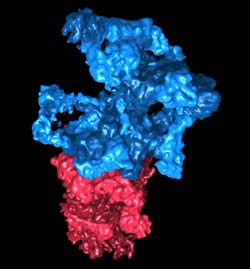
The "regulatory particle" (in blue) detects the proteins tagged with ubiquitin and prepares them for degradation. The "core particle" (in red) breaks the proteins down into their single components. Credit: Julio Ortiz / Copyright: MPI of Biochemistry<br>
Scientists at the Max Planck Institute (MPI) of Biochemistry recently succeeded in revealing the structure of the cellular protein degradation machinery (26S proteasome) by combining different methods of structural biology.
The results of collaboration with colleagues from the University of California, San Francisco and the Swiss Federal Institute of Technology Zurich (ETH Zürich) represent an important step forward in the investigation of the 26S proteasome. The findings have now been published in Proceedings of the National Academy of Sciences.
At any given point in time, cells may contain only the proteins that are needed at exactly this moment. Otherwise, undesirable reactions can occur which could cause cancer or other diseases. Furthermore, the proteins have to be folded correctly to fulfill their tasks. Misfolded proteins can clump into aggregates, and neurodegenerative diseases such as Alzheimer's or Parkinson's may be the consequence. In order to prevent this, several mechanisms in the body regulate the number of proteins in the cell and degrade proteins if necessary.
“Cellular waste disposal” – the 26S proteasome – plays an important role in protein degradation. First, misfolded and potentially dangerous proteins are tagged with molecules called ubiquitin. The 26S proteasome detects the tagged proteins and breaks them down into small fragments, which are then recycled. Scientists in the team of Wolfgang Baumeister, head of the research department “Molecular Structural Biology” at the MPI of Biochemistry, have now been able to reveal its structure.
Many puzzle pieces lead to one structure
“The structure of the 26S proteasome changes continuously,” explained Friedrich Förster, head of the research group “Modeling of Protein Complexes” at the MPI of Biochemistry. “That is why until now it could not be explained by means of traditional approaches, such as only using X-ray crystallography. We had to combine different methods to be successful.” Electron microscopy and mass spectrometry helped to reveal the general structure of the 26S proteasome. X-ray crystallography provided detailed insights into specific areas of the molecule. The researchers then used computer software to integrate the different data and generate an overall picture.
Based on these results, the researchers next want to find out how the different mechanisms of protein degradation work in detail. “We have already developed a hypothesis of how exactly the 26S proteasome detects tagged proteins and processes them,” said Stefan Bohn, scientist at the MPI of Biochemistry. The complete elucidation of the 26S proteasome and its underlying mechanisms could also be of medical importance: “Cellular waste disposal” is a therapeutic target for cancer und neurodegenerative diseases.
Media Contact
More Information:
http://www.biochem.mpg.deAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Properties of new materials for microchips
… can now be measured well. Reseachers of Delft University of Technology demonstrated measuring performance properties of ultrathin silicon membranes. Making ever smaller and more powerful chips requires new ultrathin…

Floating solar’s potential
… to support sustainable development by addressing climate, water, and energy goals holistically. A new study published this week in Nature Energy raises the potential for floating solar photovoltaics (FPV)…

Skyrmions move at record speeds
… a step towards the computing of the future. An international research team led by scientists from the CNRS1 has discovered that the magnetic nanobubbles2 known as skyrmions can be…





















