This AI birdwatcher lets you 'see' through the eyes of a machine

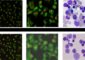
A Duke team trained a computer to identify up to 200 species of birds from just a photo. Given a photo of a mystery bird (top), the A.I. spits out heat maps showing which parts of the image are most similar to typical species features it has seen before. Credit: Chaofan Chen, Duke University
It can take years of birdwatching experience to tell one species from the next. But using an artificial intelligence technique called deep learning, Duke University researchers have trained a computer to identify up to 200 species of birds from just a photo.
The real innovation, however, is that the A.I. tool also shows its thinking, in a way that even someone who doesn't know a penguin from a puffin can understand.
The team trained their deep neural network — algorithms based on the way the brain works — by feeding it 11,788 photos of 200 bird species to learn from, ranging from swimming ducks to hovering hummingbirds.
The researchers never told the network “this is a beak” or “these are wing feathers.” Given a photo of a mystery bird, the network is able to pick out important patterns in the image and hazard a guess by comparing those patterns to typical species traits it has seen before.
Along the way it spits out a series of heat maps that essentially say: “This isn't just any warbler. It's a hooded warbler, and here are the features — like its masked head and yellow belly — that give it away.”
Duke computer science Ph.D. student Chaofan Chen and undergraduate Oscar Li led the research, along with other team members of the Prediction Analysis Lab directed by Duke professor Cynthia Rudin.
They found their neural network is able to identify the correct species up to 84% of the time — on par with some of its best-performing counterparts, which don't reveal how they are able to tell, say, one sparrow from the next.
Rudin says their project is about more than naming birds. It's about visualizing what deep neural networks are really seeing when they look at an image.
Similar technology is used to tag people on social networking sites, spot suspected criminals in surveillance cameras, and train self-driving cars to detect things like traffic lights and pedestrians.
The problem, Rudin says, is that most deep learning approaches to computer vision are notoriously opaque. Unlike traditional software, deep learning software learns from the data without being explicitly programmed. As a result, exactly how these algorithms 'think' when they classify an image isn't always clear.
Rudin and her colleagues are trying to show that A.I. doesn't have to be that way. She and her lab are designing deep learning models that explain the reasoning behind their predictions, making it clear exactly why and how they came up with their answers. When such a model makes a mistake, its built-in transparency makes it possible to see why.
For their next project, Rudin and her team are using their algorithm to classify suspicious areas in medical images like mammograms. If it works, their system won't just help doctors detect lumps, calcifications and other symptoms that could be signs of breast cancer. It will also show which parts of the mammogram it's homing in on, revealing which specific features most resemble the cancerous lesions it has seen before in other patients.
In that way, Rudin says, their network is designed to mimic the way doctors make a diagnosis. “It's case-based reasoning,” Rudin said. “We're hoping we can better explain to physicians or patients why their image was classified by the network as either malignant or benign.”
###
The team is presenting a paper on their findings at the Thirty-third Conference on Neural Information Processing Systems (NeurIPS 2019) in Vancouver on December 12.
Other authors of this study include Daniel Tao and Alina Barnett of Duke and Jonathan Su at MIT Lincoln Laboratory.
CITATION: “This Looks Like That: Deep Learning for Interpretable Image Recognition,” Chaofan Chen, Oscar Li, Daniel Tao, Alina Barnett, Jonathan Su and Cynthia Rudin. Electronic Proceedings of the Neural Information Processing Systems Conference. December 12, 2019.
Media Contact
All latest news from the category: Information Technology
Here you can find a summary of innovations in the fields of information and data processing and up-to-date developments on IT equipment and hardware.
This area covers topics such as IT services, IT architectures, IT management and telecommunications.
Newest articles

High-energy-density aqueous battery based on halogen multi-electron transfer
Traditional non-aqueous lithium-ion batteries have a high energy density, but their safety is compromised due to the flammable organic electrolytes they utilize. Aqueous batteries use water as the solvent for…

First-ever combined heart pump and pig kidney transplant
…gives new hope to patient with terminal illness. Surgeons at NYU Langone Health performed the first-ever combined mechanical heart pump and gene-edited pig kidney transplant surgery in a 54-year-old woman…

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…