Grasping the Tree of Life: There is an App for That Too

The new "TimeTree" application lets anyone with an Apple iPhone harness a vast Internet storehouse of data about the diversity of life, from bacteria to humans. The intuitive interface is designed to answer a simple question, quickly and authoritatively: how long ago did species A and species B share a common ancestor?
"Our new iPhone app can be fun for people who want to learn how long ago their cat and dog began evolving down different evolutionary paths, in addition to being a useful scientific tool," said Blair Hedges, professor of biology at Penn State University. The new smartphone app for the iPhone, iPod, and iPad gives anyone the power to explore an area at the forefront of comparative biology and to find his or her place in the timetree of life. The app is the latest extension of the Timetree of Life project, which was conceived by Hedges with Sudhir Kumar, a Ph.D. graduate of Penn State who now is director of the Biodesign Institute's Center for Evolutionary Medicine and Informatics at Arizona State University.
To download the free TimeTree app from the Apple Store, use "TimeTree" as a search term. An instructive video that briefly describes the TimeTree application for the iPhone is online at
Although the TimeTree app provides a sophisticated means of mining scientific knowledge, using it is easy. Simply type the names of two organisms — for example, swordfish and sardine — into the iPhone interface. TimeTree searches its massive web archive and returns its findings within seconds. Swordfish (Xiphias gladius) and sardine (Sardina pilchardus) shared a common ancestor some 245 million years ago, before swimming their separate ways. Along the left margin of the iPhone display is a geological timescale with data points marking each scientific study TimeTree used to reach its result. "One of the most important things about this knowledgebase," Kumar said, "is that it makes it possible for anyone to see the current agreements and disagreements in the field — immediately."
Timetrees are used widely in research in evolutionary biology, as well as in other fields. They are used to explore the tree of life with the best scientific estimates of two critical factors: the historical order in which the different lifeforms branched off in new evolutionary directions on the tree of life — the phylogeny factor — plus the number of millions of years ago when these branching events occurred — the timescale factor. Timetrees are used in the realm of human health, for example, to determine when disease-causing organisms first appeared and the speed at which their genes have changed — information that could lead to better treatments or cures.
Astrobiologists and geologists also use timetrees to study how life arose and diversified and how organisms affected the Earth's environment, including the atmosphere, throughout time.
Click on image for high-resolution file.
The Timetree of Life from an assembly of individual timetrees. Each of the 1,610 terminal branches represents a family or family-level taxon.The ultimate goal of the whole Timetree of Life project, according to Hedges, is "to chart the timescale of life — to discover when each species and all their ancestors originated, all the way back to the origin of life some four billion years ago." The first phase of this ambitious undertaking appeared last year with the simultaneous release of an online resource called "TimeTreeWeb," and an accompanying reference book, titled "The Timetree of Life" (Oxford University Press), which is edited by Hedges and Kumar and contains contributions from 105 leading scientific authorities. The TimeTree book and the web resource are freely available at www.timetree.org. The Timetree of Life web resource and iPhone app draw on The National Center for Biotechnology Information's comprehensive taxonomy browser, which contains the names and phylogenetic lineages of more than 160,000 organisms.
The resources that Hedges and Kumar have compiled in the Timetree of Life project so far form a public knowledgebase, accumulating and cataloging thousands of evolutionary-branching times for organisms available in the peer-reviewed scientific literature. TimeTree is both easier and more versatile than traditional means of searching for information on the divergence of species. One of the key advantages is TimeTree's ability to access graphical information that previously had been archived in scientific studies, inaccessible to traditional methods of data retrieval that primarily are text based. Another advantage is TimeTree's highly dynamic character — information from new scientific discoveries is being added to the knowledgebase continually, revealing the TimeTree of Life with ever-sharpening detail.
"We're still in the infancy of timetree building," Hedges said. "There are millions and millions of known species whose genes scientists worldwide are working to sequence, and there are many more species that have yet to be discovered. We are looking forward to adding these emerging new data to the TimeTree knowledgebase so that everyone can easily explore the tree of life and its timescale."
Support for developing the Timetree of Life project, including its smartphone app, has come from the U. S. National Science Foundation, the Astrobiology Institute of the U. S. National Aeronautics and Space Administration, the Science Foundation of Arizona, and the Biodesign Institute at Arizona State University.
CONTACTS
Sudhir Kumar at Arizona State: (+1) 623-225-5230, s.kumar@asu.edu
Blair Hedges at Penn State: (+1) 814-865-9991, sbh1@psu.edu
Joe Caspermeyer (PIO at Arizona State): (+1) 480-727-0369
Barbara Kennedy (PIO at Penn State): (+1) 814-863-4682, science@psu.edu
Media Contact
More Information:
http://www.psu.eduAll latest news from the category: Information Technology
Here you can find a summary of innovations in the fields of information and data processing and up-to-date developments on IT equipment and hardware.
This area covers topics such as IT services, IT architectures, IT management and telecommunications.
Newest articles
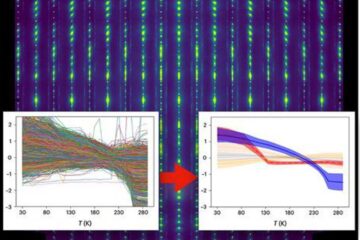
Machine learning algorithm reveals long-theorized glass phase in crystal
Scientists have found evidence of an elusive, glassy phase of matter that emerges when a crystal’s perfect internal pattern is disrupted. X-ray technology and machine learning converge to shed light…
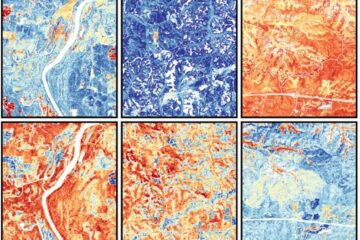
Mapping plant functional diversity from space
HKU ecologists revolutionize ecosystem monitoring with novel field-satellite integration. An international team of researchers, led by Professor Jin WU from the School of Biological Sciences at The University of Hong…

Inverters with constant full load capability
…enable an increase in the performance of electric drives. Overheating components significantly limit the performance of drivetrains in electric vehicles. Inverters in particular are subject to a high thermal load,…





















