Discovery of first demethylase molecule, a long-sought gene regulator

Could be target for cancer therapeutics
MA-Researchers have discovered an enzyme that plays an important role in controlling which genes will be turned on or off at any given time in a cell. The novel protein helps orchestrate the patterns of gene activity that determine normal cell function. Their disruption can lead to cancer.
The elusive enzyme, whose presence in cells was suspected but not proven for decades, came to light in the laboratory of Yang Shi, HMS professor of pathology, and is described in a study published in the Dec. 16 online edition of Cell and appearing in the Dec. 29 print edition.
“This discovery will have a huge impact on the field of gene regulation,” said Fred Winston, an HMS professor of genetics who was not involved with the work. “Shi and his colleagues discovered something that many people didn’t believe existed.”
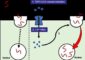
The enzyme, a histone demethylase, removes methyl groups appended to histone proteins that bind DNA and help regulate gene activity. “Previously, people thought that histone methylation was stable and irreversible,” said Shi. “The fact that we’ve identified a demethylase suggests a more dynamic process of gene regulation via methylation of histones. The idea of yin and yang is universal in biology; our results show that histone methylation is no different.”
In the cell, yarnlike strands of DNA wrap around protein scaffolds built of histones. The histones organize DNA into a packed structure that can fit into the nucleus, and the packing determines whether the genes are available to be read or not. Acetyl, methyl, or other chemical tags appended to the histones determine how the histones and DNA interact to form a chromatin structure that either promotes gene activity or represses it.
Some histone tags, particularly acetyl groups, are known to be easily added and removed, helping genes to flick on and off when needed. But the addition of methyl groups was considered a one-way process that could only be reversed by the destruction of histones and their replacement with new ones. Part of the reason scientist believed this was that no one had isolated a demethylase, despite an active search.
The Shi lab was not among those in the hunt, but they stumbled onto the demethylase while probing the function of a new gene repressor protein. Postdoctoral fellow Yujiang Shi had exhausted the likely possibilities for how the mystery protein worked to suppress gene activity, so one day he tried an unlikely experiment. He had the purified protein in a test tube and decided to feed it methylated histones. His finding, that the enzyme could efficiently chew off the methyl group, leaving behind intact, unmodified histone left the postdoc Shi shaking with excitement. “Forty years ago some scientists speculated that histone demethylases existed,” he said. “At first, I thought it was impossible that this protein was it.” After reproducing the results using several different biochemical techniques, he began to feel comfortable that they had found the first demethylase.
Their enzyme didn’t remove just any methyl group from histone. Instead, it removed a very specific methyl found on lysine 4 (K4) of histone 3 (H3). H3K4 methylation is associated with active transcription, so its removal would be consistent with the gene repression function they had identified.
Now that the first demethylase has been recognized, researchers will certainly find more. “This cannot be the only demethylase,” said Shi.
Genes turning on at the wrong time or in the wrong place is a hallmark of cancer cells. In some tumors, high levels of methylation of H3K4 seem to play a role in activating genes that drive abnormal cell growth. The discovery of this H3K4 demethylase suggests a way to counterbalance this progrowth signal in some tumors. And if previous experience with histone deacetylases is any guide, the demethylases could one day be targets for cancer therapeutics.
“These findings will impact every walk of biology,” said David Allis of Rockefeller University, a leader in studying the regulation and biological roles of histone tags. “Histone modifications are highly dynamic on-off switches that the cell throws a lot. These modifications affect everything DNA does, and getting the enzyme means you’ve got one upstream point of regulation. This will open up a wealth of new experiments.”
Media Contact
More Information:
http://hms.harvard.edu/All latest news from the category: Health and Medicine
This subject area encompasses research and studies in the field of human medicine.
Among the wide-ranging list of topics covered here are anesthesiology, anatomy, surgery, human genetics, hygiene and environmental medicine, internal medicine, neurology, pharmacology, physiology, urology and dental medicine.
Newest articles

Combatting disruptive ‘noise’ in quantum communication
In a significant milestone for quantum communication technology, an experiment has demonstrated how networks can be leveraged to combat disruptive ‘noise’ in quantum communications. The international effort led by researchers…
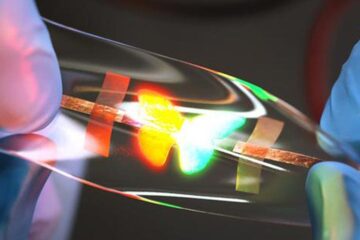
Stretchable quantum dot display
Intrinsically stretchable quantum dot-based light-emitting diodes achieved record-breaking performance. A team of South Korean scientists led by Professor KIM Dae-Hyeong of the Center for Nanoparticle Research within the Institute for…

Internet can achieve quantum speed with light saved as sound
Researchers at the University of Copenhagen’s Niels Bohr Institute have developed a new way to create quantum memory: A small drum can store data sent with light in its sonic…