Puzzle solved on how influenza builds its infectious seeds

By solving a long-standing puzzle about how the influenza virus assembles its genetic contents into infectious particles that enable the virus to spread from cell to cell, scientists have opened a new gateway to a better understanding of one of the world’s most virulent diseases.
This insight into the genetic workings that underpin infection by flu, reported today (January 27, 2003) in the Proceedings of the National Academy of Sciences (PNAS), provides not only a better basic understanding of how flu and other viruses work, but holds significant promise for new and better vaccines and drugs to combat the disease by exposing the genetic trick it uses to form virus particles.
The new work is reported by a group led by Yoshihiro Kawaoka, a professor of pathobiological sciences at the University of Wisconsin-Madison School of Veterinary Medicine, who has a joint appointment at the University of Tokyo. The group’s work describes how the flu virus selectively assembles the series of genetic subunits that make up the virus’ entire genome, which are needed to form the particles that shuttle the virus from cell to cell.
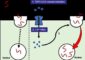
It has long been known that the influenza A virus genome requires eight different segments of RNA or ribonucleic acid to be complete. But how those RNA segments are recruited to form a complete flu genome within a cell and create particles known as virions that can go on to infect other cells was unknown.
Now, the group led by Kawaoka, in a series of detailed experiments, has uncovered a molecular signal on a single strand of RNA that is critical in the infection process. This signal selectively recruits the necessary set of eight viral RNA strands to make a complete influenza genome within the infectious particles for the influenza A strain, the most virulent and potentially dangerous flu virus.
“What’s unique about this virus is that its genome is fragmented into eight RNA segments,” Kawaoka said. “How those eight segments incorporate into a virus particle was not known.”
Viruses are masters of cellular chicanery, typically co-opting a cell’s own reproductive machinery to make copies of itself in order to infect other cells. By making artificial flu viruses with varying numbers of RNA segments, Kawaoka’s group was able to show that the virus is at its most prolific when all eight RNA segments are in place: “Our results show that when cells contain eight different segments (of viral RNA) they produce virions most efficiently.”
That finding, according to Kawaoka, indicates that the individual RNA segments each make unique contributions toward the recruitment and assembly of disparate RNA fragments into a complete influenza genome. The discovery may help pinpoint the molecular Achilles’ heel of influenza, and enable the development of new vaccines and antiviral drugs that would interfere with the ability of the virus to assemble its genome within a cell and go about the business of producing particles to infect other cells.
“These eight different segments are critical for formation of virus particles,” Kawaoka said.
Kawaoka’s work is especially important because the strain of virus used in the study, influenza A, includes the viruses responsible for the pandemics that, from time to time, sweep the globe killing millions. The most notable in recent history, the Spanish flu pandemic of 1918, killed an estimated 20 million people worldwide in a matter of months. Scientists believe it is only a matter of time before a similar pandemic recurs as the flu virus is capable of changing to avoid immune system defenses and vaccines deployed against it.
The report by Kawaoka’s team lends support to one of two competing models proposed for the generation of infectious influenza A virions. One view held that the RNA segments assembled in a random manner, depending on some common feature among the eight RNA segments to draw them randomly into the influenza genome. The selective model, which the new work supports, holds that specific structures in the individual RNA segments are responsible for combining the eight genetic fragments into a virion.
In addition to Kawaoka, members of the group publishing the report in today’s PNAS include: Yutaka Fujii of Hiroshima University, the University of Tokyo and the UW-Madison School of Veterinary Medicine; Hideo Goto and Tokiko Watanabe of the University of Tokyo and the Japan Science and Technology Corp.; and Tetsuya Yoshida of Hiroshima University.
The study was supported by the U.S. National Institute of Allergy and Infectious Diseases through a Public Health Service Research grant; the Japanese Ministry of Education, Culture, Sports, Science and Technology; and the Japan Science and Technology Corp.
Terry Devitt 608-262-8282, trdevitt@wisc.edu
Media Contact
More Information:
http://www.wisc.edu/All latest news from the category: Health and Medicine
This subject area encompasses research and studies in the field of human medicine.
Among the wide-ranging list of topics covered here are anesthesiology, anatomy, surgery, human genetics, hygiene and environmental medicine, internal medicine, neurology, pharmacology, physiology, urology and dental medicine.
Newest articles

High-energy-density aqueous battery based on halogen multi-electron transfer
Traditional non-aqueous lithium-ion batteries have a high energy density, but their safety is compromised due to the flammable organic electrolytes they utilize. Aqueous batteries use water as the solvent for…

First-ever combined heart pump and pig kidney transplant
…gives new hope to patient with terminal illness. Surgeons at NYU Langone Health performed the first-ever combined mechanical heart pump and gene-edited pig kidney transplant surgery in a 54-year-old woman…

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…