A step toward controlling Huntington's disease?

Johns Hopkins researchers have identified a natural mechanism that might one day be used to block the expression of the mutated gene known to cause Huntington’s disease. Their experiments offer not an immediate cure, but a potential new approach to stopping or even preventing the development of this relentless neurodegenerative disorder.
Huntington’s disease is a rare, fatal disorder caused by a mutation in a single gene and marked by progressive brain damage. Symptoms, which typically first appear in midlife, include jerky twitch-like movements, coordination troubles, psychiatric disorders and dementia. Although the gene responsible for Huntington’s was identified in 1993, there is no cure, and there are no treatments are available even to slow its progression.
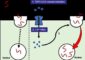
The disorder is caused by a mutation in the huntingtin gene (HTT). The mutation occurs when a section of DNA, which normally varies in length from one person to another, is too long. The result is the production of an abnormal and toxic version of the huntingtin protein. The mutation has a second unfortunate effect, the Johns Hopkins researchers discovered — it reduces a natural braking mechanism that might otherwise keep the amount of toxic huntingtin protein in check and keep the disease from developing.
“The idea of being able to harness the powers of this natural mechanism for the benefit of Huntington’s patients is a totally new way of thinking about therapy for the disease,” says Russell L. Margolis, M.D., a professor of psychiatry and behavioral sciences at the Johns Hopkins University School of Medicine and leader of the team publishing results of the study online in the journal Human Molecular Genetics.
Currently, a leading strategy among Huntington’s disease researchers is to try to suppress the expression of the mutant gene by introducing fragments of DNA meant to bind with and sabotage the ability of the gene to make the damaging protein. The goal of this approach is to prevent the mutant HTT from being expressed in the brain and potentially slow, if not stop, the disease’s march. Although cell and animal models have shown promise, Margolis and other researchers worry that getting just the right amount of DNA into the right portions of the brain may be a difficult or risky task, likely involving injections into cerebral spinal fluid or the brain itself. The feasibility of this approach remains unknown, he adds.
The new study suggests an alternative focus — manipulating the newly identified natural “brake” with a drug so that more of the brake is made, which can then specifically stop or slow production of the huntingtin protein. “Whether it’s possible to do this and do it safely remains to be seen,” Margolis says, “but this gives us another approach to explore.”
On the strand of DNA opposite the huntingtin gene, the researchers found another gene, which they named huntingtin antisense. This gene also includes the Huntington’s disease mutation. In normal brain tissue and in cells growing in the laboratory without the Huntington’s disease mutation, Margolis and his team determined that huntingtin antisense acts to inhibit the amount of huntingtin gene that is expressed. But in brain tissue and cells with the Huntington’s disease mutation, there is less huntingtin antisense gene expressed, so the biochemical foot is essentially taken off the brake, leaving a toxic amount of huntingtin protein. Reapplying the brake, by experimentally altering cells grown in culture so that they express a large amount of huntingtin antisense, decreased the amount of the toxic huntingtin protein.
Huntington’s disease was first described in the medical literature in 1872, but it wasn’t until 1993 that the gene mutation was discovered “with hopes that the discovery would quickly lead to treatment,” Margolis says. But the disease has proven unexpectedly complicated, with dozens of different pathways implicated as potential causes of cell damage and death, he adds.
People with a single copy of the mutated gene will get Huntington’s disease, which afflicts roughly 30,000 people in the United States. “It is a terrible disease in which family members can find out what’s coming and are just waiting for the symptoms to present themselves,” Margolis says. “We need to find ways to help them.”
This study was funded by the National Institutes of Health.
Other Hopkins researchers involved in the study include Daniel W. Chung, M.A.; Dobrila D. Rudnicki , Ph.D.; and Lan Yu, Ph.D.
For more information:
http://www.hopkinsmedicine.org/psychiatry/specialty_areas/huntingtons_disease/
Media Contact
More Information:
http://www.jhmi.eduAll latest news from the category: Health and Medicine
This subject area encompasses research and studies in the field of human medicine.
Among the wide-ranging list of topics covered here are anesthesiology, anatomy, surgery, human genetics, hygiene and environmental medicine, internal medicine, neurology, pharmacology, physiology, urology and dental medicine.
Newest articles

Silicon Carbide Innovation Alliance to drive industrial-scale semiconductor work
Known for its ability to withstand extreme environments and high voltages, silicon carbide (SiC) is a semiconducting material made up of silicon and carbon atoms arranged into crystals that is…

New SPECT/CT technique shows impressive biomarker identification
…offers increased access for prostate cancer patients. A novel SPECT/CT acquisition method can accurately detect radiopharmaceutical biodistribution in a convenient manner for prostate cancer patients, opening the door for more…

How 3D printers can give robots a soft touch
Soft skin coverings and touch sensors have emerged as a promising feature for robots that are both safer and more intuitive for human interaction, but they are expensive and difficult…