Splice variants reveal connections among autism genes
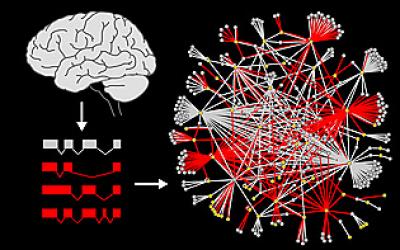
Splicing variants (red) of autism genes were cloned from the brain and screened for interactions. The image on the right represents the network of interactions. Gray lines are interactions from a single isoform; red lines are interactions from additional isoforms of autism candidate genes (yellow circles). Credit: UC San Diego School of Medicine
In their study, published in the April 11, 2014 online issue of Nature Communications, the scientists isolated hundreds of new variants of autism genes from the human brain, and then screened their protein products against thousands of other proteins to identify interacting partners. Proteins produced by alternatively-spliced autism genes and their many partners formed a biological network that produced an unprecedented view of how autism genes are connected.
“When the newly discovered splice forms of autism genes were added to the network, the total number of interactions doubled,” said principal investigator Lilia Iakoucheva, PhD, assistant professor in the Department of Psychiatry at UC San Diego. In some cases, the splice forms interacted with a completely different set of proteins. “What we see from this network is that different variants of the same protein could alter the wiring of the entire system,” Iakoucheva said.
“This is the first proteome-scale interaction network to incorporate alternative splice forms,” noted Marc Vidal, PhD, CCSB director and a co-investigator on the study. “The fact that protein variants produce such diverse patterns of interactions is exciting and quite unexpected.”
The new network also illuminated how multiple autism genes connect to one another. The scientists found that one class of mutations involved in autism, known as copy number variants, involve genes that are closely connected to each other directly or indirectly through a common partner. “This suggests that shared biological pathways may be disrupted in patients with different autism mutations,” said co-first author Guan Ning Lin, PhD, a postdoctoral fellow in Iakoucheva's laboratory.
Beyond providing greater breadth and depth around autism proteins, the network represents a new resource for future autism studies, according to Iakoucheva. For example, she said the physical collection of more than 400 splicing variants of autism candidate genes could be used by other researchers interested in studying a specific protein variant. Some of the highly connected network partners may also represent potential drug targets. All interaction data will reside in the publicly available National Database of Autism Research.
“With this assembled autism network, we can begin to investigate how newly discovered mutations from patients may disrupt this network,” said Iakoucheva. “This is an important task because the mechanism by which mutant proteins contribute to autism in 99.9 percent of cases remains unknown.”
Co-lead authors of the study are Roser Corominas, Guan Ning Lin and Shuli Kang of the Department of Psychiatry, UCSD and Xinping Yang of the CCSB, Dana-Farber Cancer Institute. Other co-authors include Yun Shen, Lila Ghamsari, Martin Broly, Maria Rodriguez, Stanley Tam, Shelly A. Trigg, Changyu Fan, Song Yi, Michael A. Calderwood, Kourosh Salehi-Ashtiani, David E. Hill and Tong Hao, CCSB, Dana-Farber Cancer Institute; Murat Tasan and Frederick P. Roth, University of Toronto; Irma Lemmens and Jan Tavernier, Ghent University; Xingyan Kuang, Nan Zhao and Dmitry Korkin, University of Missouri; Dheeraj Malholtra, Jacob J. Michaelson and Jonathan Sebat, Beyster Center for Genomics of Psychiatric Diseases and UCSD Department of Psychiatry; Vladimir Vacic, New York Genome Center; and Steve Horvath, UCLA.
This research was funded, in part, by National Institutes of Health grants R01HD065288, R01MH091350, R01HG001715 and MH076431, the Ellison Foundation, the Dana-Farber Cancer Institute Strategic Initiative, the Simons Foundation Autism Research Initiative, a Canadian Institute for Advanced Research fellowship, the Canada Excellence Research Chairs Program and the National Science Foundation.
Media Contact
More Information:
http://www.ucsd.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Bringing bio-inspired robots to life
Nebraska researcher Eric Markvicka gets NSF CAREER Award to pursue manufacture of novel materials for soft robotics and stretchable electronics. Engineers are increasingly eager to develop robots that mimic the…

Bella moths use poison to attract mates
Scientists are closer to finding out how. Pyrrolizidine alkaloids are as bitter and toxic as they are hard to pronounce. They’re produced by several different types of plants and are…

AI tool creates ‘synthetic’ images of cells
…for enhanced microscopy analysis. Observing individual cells through microscopes can reveal a range of important cell biological phenomena that frequently play a role in human diseases, but the process of…





















