Scientists determine family tree for most-endangered bird family in the world

Not only have the researchers determined the types of finches that the honeycreeper family originally evolved from, but they have also linked the timing of that rapid evolution to the formation of the four main Hawaiian Islands.
“There were once more than 55 species of these colorful songbirds, and they are so diverse that historically it wasn't even entirely clear that they were all part of the same group,” said Heather Lerner, who was a postdoctoral researcher at the Smithsonian Conservation Biology Institute's Center for Conservation and Evolutionary Genetics when she conducted this research, and is currently an assistant professor of biology at Earlham College and Joseph Moore Museum director.
“Some eat seeds, some eat fruit, some eat snails, some eat nectar. Some have the bills of parrots, others of warblers, while some are finch-like and others have straight, thin bills. So the question that we started with was how did this incredible diversity evolve over time,” Lerner said.
The answer is unique to the Hawaiian Islands, which are part of a conveyor belt of island formation, with new islands popping up as the conveyor belt moves northwest. Each island that forms represents a blank slate for evolution, so as one honeycreeper species moves from one island to a new island, those birds encounter new habitat and ecological niches that may force them to adapt and branch off into distinct species.
The researchers looked at the evolution of the Hawaiian honeycreepers after the formation of Kauai-Niihau, Oahu, Maui-Nui and Hawaii. The largest burst of evolution into new species, called a radiation, occurred between 4 million and 2.5 million years ago, after Kauai-Niihau and Oahu formed but before the remaining two large islands existed, and resulted in the evolution of six of 10 distinct groups of species characterized by different sizes, shapes and colors.
“This radiation is one of the natural scientific treasures that the archipelago offers out in the middle of the Pacific,” said Helen James, a research zoologist at the Smithsonian's National Museum of Natural History and a co-author of the paper. “It was fascinating to be able to tie a biological system to geological formation and allowed us to become the first to offer a full picture of these birds' adaptive history.”
James' previous work on Hawaiian birds' morphology, the branch of biology that deals with form and structure of organisms, played a pivotal role in determining which avian species to survey to determine the closest living relatives of the Hawaiian honeycreepers.
Using genetic data from 28 bird species that seemed similar to the honeycreepers morphologically, genetically or that shared geographic proximity, the paper's authors determined that the various honeycreeper species evolved from Eurasian rosefinches. Unlike most other ancestral bird species that came from North America and colonized the Hawaiian Islands, the rosefinch likely came from Asia, the scientists found.
“There is a perception that there are no species remaining that are actually native to Hawaii, but these are truly native birds that are scientifically valuable and play an important and unique ecological function,” said Rob Fleischer, head of SCBI's Center for Conservation and Evolutionary Genetics and a co-author of the paper.
Fleischer has been studying the genetics, evolution and conservation of these birds for more than 25 years. “I'm thrilled that we finally had enough DNA sequence and the necessary technology to become the first to produce this accurate and reliable evolutionary tree.”
The diversity of Hawaiian honeycreepers has taken a huge hit, with more than half of the known 56 species already extinct. The paper's researchers focused on the 19—now 18—species that have not gone extinct, but of those, six are considered critically endangered by the International Union for Conservation of Nature, four are considered endangered and five are vulnerable.
The next, ongoing step in the research is to use museum specimens and subfossil bones to determine where the extinct species fit into the evolutionary family tree, or phylogeny, to see if the new lineages fit into the overall pattern found in this study.
To analyze the DNA for the study that came out this week, the researchers used specialized next generation sequencing protocols developed by Michi Hofreiter's group at the Max Planck Institute for Evolutionary Anthropology. To look at ancient DNA that is by nature damaged or degraded, they are using additional innovative techniques to capture the DNA and come up with a sufficiently informative dataset.
The study's authors from SCBI are Lerner and Fleischer. The additional authors are James from the Smithsonian's National Museum of Natural History, Hofreiter from the University of York and Matthias Meyer from the Max Planck Institute for Evolutionary Anthropology. The work was funded by the National Science Foundation.
The Smithsonian Conservation Biology Institute plays a key role in the Smithsonian's global efforts to understand and conserve species and train future generations of conservationists. Headquartered in Front Royal, Va., SCBI facilitates and promotes research programs based at Front Royal, the National Zoo in Washington, D.C., and at field research stations and training sites worldwide.
These findings will be published in the hard-copy version of Current Biology Nov. 8, with Lerner as lead author. (A PDF version of the paper is available online on Current Biology's media pages.)
Media Contact
More Information:
http://www.si.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
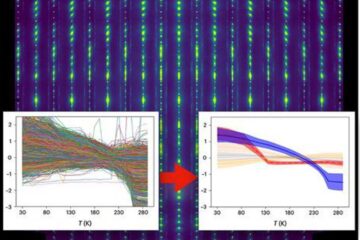
Machine learning algorithm reveals long-theorized glass phase in crystal
Scientists have found evidence of an elusive, glassy phase of matter that emerges when a crystal’s perfect internal pattern is disrupted. X-ray technology and machine learning converge to shed light…
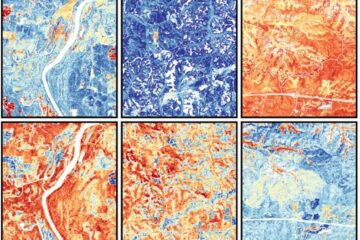
Mapping plant functional diversity from space
HKU ecologists revolutionize ecosystem monitoring with novel field-satellite integration. An international team of researchers, led by Professor Jin WU from the School of Biological Sciences at The University of Hong…

Inverters with constant full load capability
…enable an increase in the performance of electric drives. Overheating components significantly limit the performance of drivetrains in electric vehicles. Inverters in particular are subject to a high thermal load,…





















