Researchers identify biochemical functions for most of the human genome

Only about 1 percent of the human genome contains gene regions that code for proteins, raising the question of what the rest of the DNA is doing. Scientists have now begun to discover the answer: About 80 percent of the genome is biochemically active, and likely involved in regulating the expression of nearby genes, according to a study from a large international team of researchers.
The consortium, known as ENCODE (which stands for “Encyclopedia of DNA Elements”), includes hundreds of scientists from several dozen labs around the world. Using genetic sequencing data from 140 types of cells, the researchers were able to identify thousands of DNA regions that help fine-tune genes’ activity and influence which genes are expressed in different kinds of cells.
Just as the sequencing of the human genome helped scientists learn how mutations in protein-coding genes can lead to disease, the new map of noncoding regions should provide some answers on how mutations in the regulatory elements lead to diseases such as lupus and diabetes, says Manolis Kellis, an associate professor of computer science at MIT, an associate member of the Broad Institute and an author of a paper describing the findings in the Sept. 5 online edition of Nature.
“Humans are 99.9 percent identical to each other, and you only have one difference in every 300 to 1,000 nucleotides,” Kellis says. “What ENCODE allows you to do is provide an annotation of what each nucleotide of the genome does, so that when it’s mutated, we can make some predictions about the consequences of the mutation.”
Kellis, who leads MIT’s Computational Biology Group, is one of the principal investigators involved in the Nature paper. The ENCODE collaboration is publishing about two dozen additional papers this week detailing the new results.
Mapping noncoding DNA
ENCODE was established in 2003 to extend our understanding of the human genome beyond protein-coding genes. One way to do that is by studying the chemical modifications of individual stretches of DNA, which control when genetic regions will be active. These modifications vary by cell type and can modify either DNA directly or the histone proteins that DNA wraps around.
To map these modifications, known collectively as the epigenome, the research groups had to collect many different kinds of data from different cell types. Some labs measured DNA or histone modifications, while others gauged the accessibility of different stretches of DNA by cutting it into fragments with enzymes.
Kellis and his group were among the computational scientists leading the effort to analyze and integrate the huge amount of data generated by different labs. “Given that we were getting more than 1,000 data sets, we had to figure out ways to automatically calibrate experiments,” says Anshul Kundaje, a research scientist in MIT’s Computational Biology Group. “We developed an almost purely automated system that did all of this.”
The ENCODE researchers found that 80 percent of the genome experiences some kind of biochemical event, such as binding to proteins that regulate how often a neighboring gene is utilized. They also discovered that the same regulatory region can play different roles, depending on what type of cell it’s acting in.
The findings should have a major impact on scientists’ understanding of human biology and how genomic variations can cause disease, says Ben Raphael, an associate professor of computer science at Brown University.
“The most exciting part is now we’re getting a whole genome annotation of functional elements,” says Raphael, who was not part of the research team. “Every time you want to understand what a particular piece of the genome is doing, you can use the data from this project.”
Human variation
The researchers also studied the conservation of nucleotides — the A, T, C and G “letters” of DNA — in the newly identified regulatory regions. Nucleotides are conserved if they remain the same over long evolutionary periods, which can be measured by analyzing the variability between species, or among individuals within a species.
A recent paper by Kellis and colleagues showed that 5 percent of noncoding DNA is conserved across mammals. In one of the ENCODE companion papers appearing online Sept. 5 in Science, Kellis and MIT postdoc Lucas Ward show that an additional 4 percent is conserved within the human lineage, suggesting that those elements control recently evolved traits, some of which are unique to humans.
When the researchers looked at the functions of genes near newly evolved regulatory regions, they found many genes that encode regulators that activate other genes. “Genes involved in the nerve growth pathway and color vision, both of which have been hypothesized to be recent innovations in the primate lineage, are enriched in human-constrained elements in non-conserved regions,” Ward says.
The researchers found that the most highly conserved nucleotides were also the ones most likely to be associated with disease when mutated. They also showed that variants associated with autoimmune diseases such as lupus and rheumatoid arthritis are located in regions active only in immune cells, while variants linked to metabolic diseases are in regions active only in liver cells.
In their next phase, the ENCODE researchers hope to determine just how those variations lead to human disease.
“What we’ve done over this series of papers is effectively paint a set of reference annotations of common human genome function,” Kellis says. “Our next steps will be to personalize these maps — to basically ask how they vary naturally between individuals, by profiling different cell types from different people, and how their variation relates to human disease and complex human traits.”
In one follow-up project, Kellis and colleagues are comparing activity levels of regulatory elements in different cell types from the same person, across many individuals. Another project is looking at DNA modification patterns across the entire genome of many individuals, in hopes of identifying how variation of specific elements relates to disease.
The research was funded by the National Human Genome Research Institute.
Media Contact
More Information:
http://www.mit.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
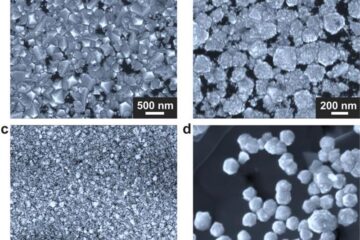
Making diamonds at ambient pressure
Scientists develop novel liquid metal alloy system to synthesize diamond under moderate conditions. Did you know that 99% of synthetic diamonds are currently produced using high-pressure and high-temperature (HPHT) methods?[2]…

Eruption of mega-magnetic star lights up nearby galaxy
Thanks to ESA satellites, an international team including UNIGE researchers has detected a giant eruption coming from a magnetar, an extremely magnetic neutron star. While ESA’s satellite INTEGRAL was observing…
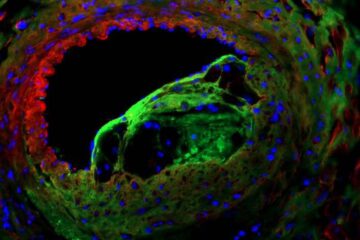
Solving the riddle of the sphingolipids in coronary artery disease
Weill Cornell Medicine investigators have uncovered a way to unleash in blood vessels the protective effects of a type of fat-related molecule known as a sphingolipid, suggesting a promising new…





















