Novel laboratory technique nudges genes into activity

The gene-activating method, which is being developed by UT Southwestern scientists, also is providing researchers with a novel research tool to investigate the role that genes play in human health.
In a paper appearing online at Nature Chemical Biology and in an upcoming edition of the journal, lead author Dr. Bethany Janowski, assistant professor of pharmacology at UT Southwestern, and her colleagues describe how they activated certain genes in cultured cells using strands of RNA to perturb the delicately balanced mixture of proteins that surround chromosomal DNA, proteins that control whether genes are turned on or off.
Dr. David Corey, professor of pharmacology and the paper’s senior author, said the results are significant because they demonstrate the most effective and consistent method to date for coaxing genes into making the proteins that carry out all of life’s functions – a process formally called gene expression.
In any medical specialty, Dr. Janowski said, there are conditions where increased gene expression would prove beneficial.
“In some disease states, it’s not that gene expression is completely turned off, but rather, the levels of expression are lower than they should be,” she said. As a result, there is an inadequate amount of a particular protein in the body. “If we can bring the level up a few notches, we might actually treat or cure the disease,” Dr. Janowski said.
For example, some genes are natural tumor suppressors, and using this method to selectively activate those genes might help the body fend off cancer, Dr. Janowski said.
Genes are segments of DNA housed in chromosomes in the nucleus of every cell and they carry instructions for making proteins. Faulty or mutated genes lead to malfunctioning, missing or over-abundant proteins, and any of those conditions can result in disease.
Surrounding the chromosome is a cloud of proteins that helps determine whether or not a particular gene’s instructions are “read” and “copied” to strands of messenger RNA, which then ferry the plans to protein-making “factories” in the cell.
In its experiments, the UT Southwestern team used strands of RNA that were tailor-made to complement the DNA sequence of a specific gene in isolated breast cancer cells. Once the RNA was introduced into the protein mix, the gene was activated, ultimately resulting in a reduced rate of growth in the cancer cells.
Dr. Corey said that while it’s clear the activating effects of the new technique are occurring at the chromosome level, and not at the messenger RNA level, more research is needed to understand the exact mechanism.
Although the RNA strands the researchers introduced – dubbed antigene RNA – were manufactured, Dr. Corey said the process by which they interact with the chromosome appears to mimic what naturally happens in the body.
“One of the reasons why these synthetic strands work so well is that we’re just adapting a natural mechanism to help deliver a man-made molecule,” Dr. Corey said. “We’re working with nature, rather than against it.”
Drs. Corey’s and Janowski’s current results are built on previous work, published in 2005 in Nature Chemical Biology, in which they found that RNA strands could turn off gene expression at the chromosome level.
The new UT Southwestern research, coupled with that from 2005, demonstrates a shift away from conventional thinking about how gene expression is naturally controlled, as well as how scientists might be able to exploit the process to develop new drug targets, Dr. Corey said.
For example, current methods to block gene expression, such as RNA interference, rely on using RNA strands to intercept and bind with messenger RNA. While RNA interference is an effective tool for studying gene expression, Dr. Janowski said, it’s more efficient to use RNA to control both activation and de-activation at the level of the chromosome.
“It goes right to the source, right to the faucet to turn the genes on or off,” she said.
Dr. Corey said many researchers have the ingrained idea that RNA only targets other RNA – such as what occurs when messenger RNA is targeted during RNA interference. “That’s what everyone is familiar with,” he said. “But the idea of RNA being used as a sort of nucleic acid modulator of chromosomes, at the level of the chromosome itself, is novel and unexpected, and it’s going to take some getting used to.”
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
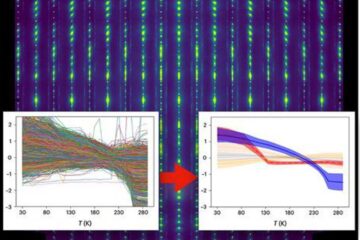
Machine learning algorithm reveals long-theorized glass phase in crystal
Scientists have found evidence of an elusive, glassy phase of matter that emerges when a crystal’s perfect internal pattern is disrupted. X-ray technology and machine learning converge to shed light…
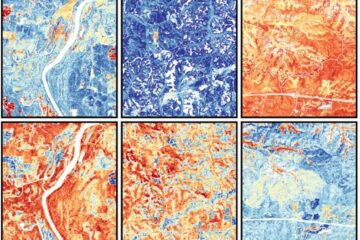
Mapping plant functional diversity from space
HKU ecologists revolutionize ecosystem monitoring with novel field-satellite integration. An international team of researchers, led by Professor Jin WU from the School of Biological Sciences at The University of Hong…

Inverters with constant full load capability
…enable an increase in the performance of electric drives. Overheating components significantly limit the performance of drivetrains in electric vehicles. Inverters in particular are subject to a high thermal load,…





















