Common Ancestry of Bacterium and Plants Could be Key to an Effective New Treatment for Chlamydia

“The unique connection between the Chlamydia bacterium and plants had been proposed by others,” said Thomas Leustek,” a professor in the department of plant biology and pathology at Rutgers' School of Environmental and Biological Sciences (formerly Cook College). “But we have now described a specific example demonstrating the common heritage. That specific example, an enzyme that supports protein production, could lead to antibiotics specific for this form of STD.”
The discovery is an unexpected turn in solving the mystery of how plants produce lysine, one of the 20 amino acids normally found in proteins. Scientists have known the specific pathways of lysine production in bacteria for more than a half-century. They also have known some of the steps by which lysine is produced in plants, but they didn’t really have the full picture. Leustek and Andre Hudson, a postdoc working in Leustek’s lab in Rutgers’ Biotechnology Center for Agriculture and the Environment, were able to solve the pathway when they discovered the gene encoding the enzyme L,L-diaminopimelate aminotransferase from the plant Arabdiopsis thaliana. The results of this discovery were published in the journal Plant Physiology in January 2006.
The gene that Leustek and Hudson had discovered was unmistakably similar to a sequence that Anthony Maurelli of the Uniformed Services University of the Health Sciences in Bethesda, Md., had detected in Chlamydia. “Further experimentation confirmed that the Chlamydial gene had the same function as the Arabidopisis gene demonstrating their common ancestry,” said Leustek. “If they evolved separately, it would be impossible for the sequences to match so closely.”
The ability to easily compare plants and bacteria is the result of genome sequencing, which has decoded the complete genetic blueprint for entire species. “This would not have been possible 10 years ago,” said Leustek. “But now we have access to more that 500 different genomes in a data base. After having identified a gene in plants, I can quickly identify the homologous gene from any bacteria in the database. As a plant biologist I wouldn’t have ever imagined that I would be working with Chlamydia. Yet, with the help of genomics I found myself working with a collaborator and publishing a paper in that area.”
Their experiments revealed that in addition to sharing genome sequences, Chlamydia and plants share similar functions as well. Furthermore, they found that the pathway used by plants to produce lysine is probably used by Chlamydia to synthesize a chemical found in bacterial cell walls. It is the synthesis of cell walls that is inhibited by penicillin. This discovery points to the likelihood that, if researchers could find an inhibitor for L,L-diaminopimelate aminotransferase they would have a new antibiotic that would target Chlamydia.
Chlamydia trachomatis is a bacteria that is responsible for a common STD. If untreated, Chlamydia infections can damage a woman's reproductive organs and lead to infertility. An estimated 2.8 million men and women in the U.S. are infected with chlamydia each year. Chlamydia can be easily treated and cured with antibiotics. However, bacteria often develop resistance to antibiotics, meaning that new ones must be continually discovered. Moreover, an inhibitor to L,L-diaminopimelate aminotransferase would be very specific for Chlamydia since this enzyme has not been found in any bacteria that live with humans.
So the hunt for a new antibiotic is on. Leustek is going to start screening for chemicals that block the enzyme. He is also using the results of his research to work on another approach, which is to characterize the structure of the enzyme so that he could design an antibiotic that would disable the pathway. This approach is somewhat like designing a key to fit a lock by opening the lock and looking inside.
The research is being done in collaboration with Charles Gilvarg from Princeton University. “He’s the biochemist who characterized the lysine pathway back in the 1950s, and so he had intimate knowledge about the steps of the pathway,” said Leustek. “And he’s the one that alerted us to the fact that plants do it differently. This is still the case, with the exception of the Chlamydia bacterium.”
The latest work, which describes the similarities in the genetic sequences of Chlamydia and plants, will be published in the Proceedings of the National Academy of Sciences’ Online Early Edition the week of November 6, 2006. In addition to Leustek, Hudson, Maurelli and Gilvarg, authors include Andrea McCoy and Nancy Adams of the Uniformed Services University of the Health Sciences.
Contact:
Michele Hujber
732-932-7000 x 4204
E-mail: hujber@aesop.rutgers.edu
Media Contact
More Information:
http://www.rutgers.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
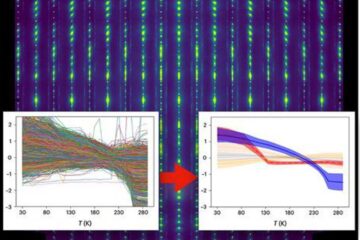
Machine learning algorithm reveals long-theorized glass phase in crystal
Scientists have found evidence of an elusive, glassy phase of matter that emerges when a crystal’s perfect internal pattern is disrupted. X-ray technology and machine learning converge to shed light…
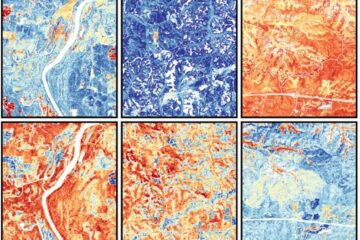
Mapping plant functional diversity from space
HKU ecologists revolutionize ecosystem monitoring with novel field-satellite integration. An international team of researchers, led by Professor Jin WU from the School of Biological Sciences at The University of Hong…

Inverters with constant full load capability
…enable an increase in the performance of electric drives. Overheating components significantly limit the performance of drivetrains in electric vehicles. Inverters in particular are subject to a high thermal load,…





















