New cell imaging method identifies aggressive cancer cells early
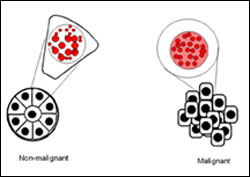
Learning about the changes in location of proteins within cell nuclei may help researchers determine the types and virulence of cancer cells and find new treatments. Purdue researcher Sophie Lelièvre and her colleagues at Lawrence Berkeley National Laboratory studied the protein NuMA in breast tissue to develop an automated technique to track, map and analyze the protein’s redistribution in different cell types. The illustration compares NuMA (in red) in the nucleus of a normal breast tissue cell (top left) to the nucleus of a cell belonging to an invasive breast tumor (top right). The drawings in black show nuclei within the organization of non-malignant and malignant cells of a normal glandular breast tissue (bottom left) and of a tumor nodule (bottom right), respectively. (Illustration by Sophie Lelièvre)
Fluorescence that illuminates a specific protein within a cell’s nucleus may be a key to identifying cancer virulence and to developing individualized treatment, according to researchers at Purdue University and Lawrence Berkeley National Laboratory.
The scientists created a technique that automatically locates and maps proteins involved in regulating cell behavior, said Sophie Lelièvre, Purdue assistant professor of basic medical sciences. The research results have for the first time made it possible to verify the distinction between multiplying cells that are harmless and those that are malignant.
Lelièvre and co-corresponding author on the study, David Knowles of the national lab, used human mammary cells to analyze nuclear protein distribution that shifted depending on whether a cell was malignant, had not yet developed a specific function or was a normally functioning mature mammary cell.
“When you look at cells that don’t yet have a specific function – aren’t differentiated, compared to fully differentiated cells, which are now capable of functioning as breast cells – the organization of proteins in the nucleus varies tremendously,” Lelièvre said. “Then looking at how the proteins in malignant cells are distributed, it’s a totally different pattern compared to normal differentiated cells.”
The research team’s study on the imaging technique and its use in 3-D mapping and analysis of nuclear protein distribution is published this week online in Proceedings of the National Academy of Sciences. Ultimately, the scientists want to use the technique to determine not only if a lesion is malignant but also the exact kind of cancer, how likely it is to spread and the most appropriate treatment for a particular patient.
“The major problem exists in the pre-malignant stages of abnormal cells in determining whether cancer will develop, what type and how invasive it will be,” Lelièvre said. “The decision then is whether to treat or not to treat and how to proceed in these preliminary stages because only a certain percentage of these patients will ultimately develop cancer.
“We want to use this technique to identify subtypes of cells within lesions that potentially could become more aggressive forms of cancer.”
Lelièvre, Knowles and their team used an antibody attached to a fluorescent molecule that targeted and linked with a specific nuclear protein from mammary tissue. When malfunctioning, this protein, named nuclear mitotic apparatus protein (NuMA), has been linked to leukemia and breast cancer.
The imaging technique the researchers developed to identify NuMA location shifts is called an automated local bright feature image analysis. It recorded the average amount of luminescence throughout the nucleus and then located the brightest spots, which were the protein. The system then automatically measured the differences in the protein’s distribution in each cell type and mapped it. This enabled the researchers to verify the changes exhibited by non-differentiated cells that were still multiplying, normal mammary cells and multiplying malignant cells.
The ability to see the protein patterns in the nucleus gives scientists one more tool in advancing the identification of types of cancer and appropriate treatment, Lelièvre said. The imaging tool should work for mapping and analyzing locations of any nuclear protein.
“We have genomics and proteomics that tell us about where genes are, whether they are functioning and interactions of genes with proteins, but no one had focused on the changing distribution of nuclear proteins,” she said. “Looking at the location of the proteins is a third part of studying cancer.
“We call it architectural proteomics because the proteins are still there but the location changes.”
These protein shifts in the nucleus also may change the protein function, Lelièvre said. The new technique to map protein location will help determine this as well. In the case of malignant cells, it may reveal what signaling process went awry causing abnormal cell growth.
“It’s as if, instead of losing an arm, your arm was placed in another location. It’s abnormal, but you have everything you need – just not in the right place,” she said. “It’s what happens in cancer, too; the needed proteins are still there but not in the right place anymore, so their function is altered.”
The misplaced proteins in their new locations change how the cell behaves and participate in the promotion of cancer, she said. Being able to measure the protein location shifts to aid in determining their function in cancer cell development will allow scientists to use the proteins as treatment targets.
“With our new system, we now will be able to look at individual cells and nuclei and possibly identify some classes of cells that could be more dangerous than others,” Lelièvre said.
The other researchers on the study were Carol Bator-Kelly, Purdue Department of Biological Science, and Damir Sudar and Mina Bissell, at the National Laboratory’s Life Sciences Division Biophysics and Cancer Biology departments.
Lelièvre is also a member of Purdue’s National Cancer Institute-designated Cancer Center and Purdue’s Oncological Science Center at Discovery Park.
Funding for this research came from the Department of Defense Breast Cancer Research Program, the Department of Energy Office of Health and Environmental Research, the Walther Cancer Institute, “Friends you Can Count On” and the Purdue University Research Foundation.
Writer: Susan A. Steeves, (765) 496-7481, ssteeves@purdue.edu
Source: Sophie Lelièvre, (765)-496-7793, lelièvre@purdue.edu
Ag Communications: (765) 494-2722; Beth Forbes, forbes@purdue.edu
Agriculture News Page
Media Contact
More Information:
http://www.purdue.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Bringing bio-inspired robots to life
Nebraska researcher Eric Markvicka gets NSF CAREER Award to pursue manufacture of novel materials for soft robotics and stretchable electronics. Engineers are increasingly eager to develop robots that mimic the…

Bella moths use poison to attract mates
Scientists are closer to finding out how. Pyrrolizidine alkaloids are as bitter and toxic as they are hard to pronounce. They’re produced by several different types of plants and are…

AI tool creates ‘synthetic’ images of cells
…for enhanced microscopy analysis. Observing individual cells through microscopes can reveal a range of important cell biological phenomena that frequently play a role in human diseases, but the process of…





















