Cellular dumping site is not garbage after all
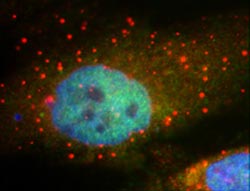
This image shows P-bodies, in red, surrounding the nucleus of a human tumor cell. The red color indicates that the P-bodies contain the protein RCK. Photo credit: John Bloom.
Cells can reuse the chemical messengers that carry genetic information to the machinery that makes proteins. Sometimes cells shuttle the messengers to storage and later reactivate them to make proteins, according to new research.
Learning how cells regulate the newly discovered “mRNA cycle” may provide insights into how the cellular machinery runs amok in diseases like cancer.
Scientists had previously thought the messenger molecules, known as mRNAs, were manufactured, used, decommissioned and then sent on a one-way journey to the garbage dump.
These cellular garbage dumps, called P-bodies, turn out to be storage depots, not landfills. After use, mRNA molecules are temporarily deactivated for storage purposes. The cell can then either destroy the mRNA or recondition pre-used mRNA so it can be put back into service if needed.
P-bodies are also involved in determining whether specific mRNAs are used to make proteins, a process called translation.
“We were surprised to find that the P-bodies were involved in regulating translation,” said research team leader Roy Parker, a Regents’ Professor of molecular and cellular biology at The University of Arizona in Tucson and an Investigator with the Howard Hughes Medical Institute. In 2003, his lab was the first to name and describe a function for P-bodies.
Parker said of the new finding, “It suggests P-bodies have a much broader role in controlling the activities of the cell than we realized.”
Parker and first author Jeff Coller report P-bodies’ role in the control of translation in the Sept. 23 issue of the journal Cell. Coller, who did the research while at UA as a postdoctoral fellow with the Howard Hughes Medical Institute, is now an assistant professor in the Center for RNA Molecular Biology at Case Western Reserve University in Cleveland, Ohio.
The Parker lab’s findings about P-bodies serving as storage depots was released online Sept. 1, 2005 and will be published in an upcoming issue of Science. Complete citations for the two papers can be found at the end of this release. The Howard Hughes Medical Institute and the National Institutes of Health funded the research.
To live and grow, cells convert the genetic instructions stored in DNA into proteins. However, only some of the myriad instructions stored in genes are useful at any one time. Researchers want to figure out how cells switch from manufacturing one type of protein to another. mRNA molecules are key in the manufacturing process because they carry the protein-assembly instructions from the DNA to the assembly plant.
Although P-bodies were initially identified as just garbage dumps for used mRNA, Parker and his colleagues suspected P-bodies played a more important role in determining which proteins a cell makes.
The researchers investigated whether two proteins known to decommission mRNA, Dhh1p and Pat1p, were involved in regulating the translation of mRNA’s instructions into proteins. The scientists did their experiments with common baker’s yeast, a one-celled organism known to scientists as Saccharomyces cerevisiae.
To see what happened if the cellular machinery didn’t work right, the researchers compared the behavior of mutant yeast cells to normal yeast cells.
Coller and Parker tested mutant cells that lacked one or both proteins to see how they compared with normal cells. Under the microscope, only 10 percent of the mutant cells that lacked both proteins had P-bodies, whereas almost all the normal cells had P-bodies. In addition, the mutant cells could no longer turn off the use of mRNAs under the appropriate conditions. Parker said, “Cells missing these proteins could no longer turn off mRNAs and could no longer make P-bodies.”
The team then engineered yeast cells to produce an overabundance of the proteins and repeated the experiments with those cells. Those cells stopped growing. Parker said, “The mRNAs are all driven away from the assembly factories. When you look through the microscope, the cells have huge P-bodies. It’s very dramatic.”
Overabundance of a human protein similar to Dhh1p occurs in many tumors, but the function of the protein is unknown, Parker said. In another experiment, the team put some mRNA, some protein-assembly plants and some of the human protein, referred to as RCK, into test tubes.
When RCK was added, mRNAs did not enter the assembly plants, suggesting that the human protein prevents cells from translating mRNA’s instructions into the proteins that cells need to thrive. “Adding this protein would screw it up,” Parker said. “The protein suppresses the translation process.”
People initially thought that switching from making one protein to another happened solely by blocking the assembly of the protein-making machinery. Coller and Parker’s Cell paper shows that the regulation occurs by determining whether or not mRNA will enter the assembly plant or be shipped to a storage depot. Contrary to previous beliefs, the team suspects the cell can bypass the assembly plant altogether and send unneeded mRNAs straight to P-bodies to be degraded or eventually recycled.
Parker said his lab’s next step is examining the potential role of P-bodies in memory and in viral infections.
Media Contact
More Information:
http://www.arizona.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Superradiant atoms could push the boundaries of how precisely time can be measured
Superradiant atoms can help us measure time more precisely than ever. In a new study, researchers from the University of Copenhagen present a new method for measuring the time interval,…

Ion thermoelectric conversion devices for near room temperature
The electrode sheet of the thermoelectric device consists of ionic hydrogel, which is sandwiched between the electrodes to form, and the Prussian blue on the electrode undergoes a redox reaction…

Zap Energy achieves 37-million-degree temperatures in a compact device
New publication reports record electron temperatures for a small-scale, sheared-flow-stabilized Z-pinch fusion device. In the nine decades since humans first produced fusion reactions, only a few fusion technologies have demonstrated…





















