UCSD discovery may provide novel method to generate medically useful proteins
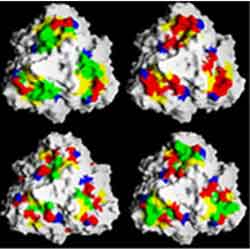
Graphic shows molecular structure of predator protein variants (colors reveal different amino acids) Credit: Jason Miller, UCSD
A team led by UCSD biochemists has discovered the mechanism by which a simple organism can produce 10 trillion varieties of a single protein, a finding that provides a new tool to develop novel drugs.
In the September 18 advance on-line publication of the journal Nature Structural and Molecular Biology, the researchers describe the mechanism by which a virus that infects bacteria—called a bacteriophage, or phage—can generate a kaleidoscope of variants of a particular protein. The paper will appear in print in Nature Structural and Molecular Biology in October.
Since this degree of protein diversity is extremely rare, recreating the process in a test tube could give researchers a new way to generate therapeutic enzymes, vaccines and other medically important proteins.
“This is only the second type of massively variable protein ever discovered,” explained Partho Ghosh, a professor of chemistry and biochemistry at UCSD who headed the research team. “Only antibodies have more variation than this protein in phage. However, the genetic mechanism used by the phage to generate this diversity is completely different from that used by animals to produce antibodies, and has the advantage of giving the protein greater stability.”
“If we can learn from these organisms how to set up a system that churns out proteins with enormous variability, it may be possible to target these new proteins to specific cells to treat disease,” said Stephen McMahon, a former postdoctoral fellow in Ghosh’s lab who conducted much of the research. “This idea has already been picked up by the biotech industry.”
The function of the massively variable phage protein is to tether the phage to the bacteria they infect. The phage “predator” protein fits into a “prey” protein on the bacteria like a three-dimensional puzzle piece. However, the bacteria are constantly changing the proteins on their surface. To keep up with the unpredictable changes in the prey protein, the phage must generate many different predator proteins for at least one to have an acceptable fit.
In their paper, the researchers describe how by altering the amino acids at one or more of just 12 sites on the predator protein, the phage are able to generate 10 trillion proteins, each with the potential to bind to a different prey protein. This variability arises as DNA is being copied into the RNA blueprint for the protein. The sequence of DNA bases at the 12 sites has unique characteristics that cause frequent mistakes to be made in the copying process. As a result, the RNA ends up specifying a different amino acid, and a protein with different structural and chemical properties is created.
Antibodies are another type predator protein that must respond to rapidly evolving prey proteins, because microorganisms are constantly altering proteins on their surfaces to evade the immune system. Unlike the phage protein, antibodies have a complicated loop structure. The size of the loops varies in addition to the amino acid building blocks that constitute the antibody protein. Although this mechanism can generate more than 100 trillion different antibodies, the researchers say replicating it in a test tube would be very challenging because the loops would have the tendency to fold incorrectly.
“Because of its stability, the phage protein makes a better model to create protein diversity in a test tube,” explained Jason Miller, a graduate student in Ghosh’s lab who conducted much of the research. “Our discovery shows that nature has provided at least two completely different methods to generate a huge amount of protein variability, and it opens up a whole new platform for protein development.”
Other contributors to the paper were Jeffrey Lawton, Department of Chemistry, Eastern University; Donald Kerkow, The Scripps Research Institute; Marc Marti-Renom, Eswar Narayanan, and Andrej Sali, Departments of Biopharmaceutical Sciences and Pharmaceutical Chemistry, University of California, San Francisco; Asher Hodes, and Jeffrey Miller, Department of Microbiology, Immunology, and Molecular Genetics, David Geffen School of Medicine and the Molecular Biology Institute, University of California, Los Angeles; and Sergei Doulatov, Department of Microbiology and Medical Genetics, University of Toronto.
Stephen McMahon is now at the Centre for Biomolecular Sciences at The University of St. Andrews in Scotland.
This research was supported by a W.M. Keck Distinguished Young Scholars in Medicine Award and a UC Discovery Grant.
Media Contact
More Information:
http://www.ucsd.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Bringing bio-inspired robots to life
Nebraska researcher Eric Markvicka gets NSF CAREER Award to pursue manufacture of novel materials for soft robotics and stretchable electronics. Engineers are increasingly eager to develop robots that mimic the…

Bella moths use poison to attract mates
Scientists are closer to finding out how. Pyrrolizidine alkaloids are as bitter and toxic as they are hard to pronounce. They’re produced by several different types of plants and are…

AI tool creates ‘synthetic’ images of cells
…for enhanced microscopy analysis. Observing individual cells through microscopes can reveal a range of important cell biological phenomena that frequently play a role in human diseases, but the process of…





















