Researchers Reveal Secret of Key Protein in Brain and Heart Function
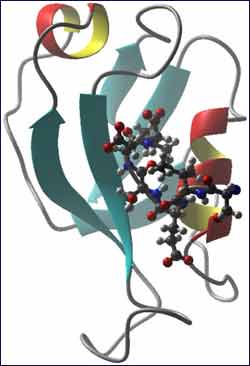
Synapse-associated protein 97. The model shows the PDZ domain of the SAP97 protein in a ribbon format, highlighting its structural elements. The intra-cellular portion of the NMDA receptor is shown as ball-and-stick format atoms. SAP97 is a scaffolding protein, facilitating nerve signals.
Brown University biologists have solved the structure of a critical piece of synapse-associated protein 97 (SAP97) found in abundance in the heart and head, where it is believed to play a role in everything from cardiac contractions to memory creation. Results are published in The Journal of Biological Chemistry.
Dale Mierke, associate professor of medical science at Brown, said that knowing how a piece of SAP97 is built is an important step. Now that part of the protein’s structure is solved, scientists can create a molecule to disable it. That, in turn, will allow them to fully understand SAP97’s role in the body. And that will point drug makers to targets for developing new ways to treat cardiac or neurological diseases.
“To arrive at a solution, you need to understand the problem,” Mierke said. “Solving protein structures opens doors for effective treatments.”
SAP97 is found mainly in the central nervous system and is known as a “scaffolding” protein. In this role, it serves as a sort of tether, grabbing proteins inside the cell critical to nerve signaling and keeping them close to N-methyl-D-asparate (NMDA) receptors at the cell surface. NMDA receptors help usher in a neurotransmitter called glutamate that is essential for learning and memory and also plays a role in drug addiction. A similar scaffolding mechanism is at work in the heart, where it affects basic functions, including the heartbeat.
SAP97 is a complex protein made up of five “domains” similar to a train comprised of an engine and four boxcars. In their experiments, Mierke, graduate student Lei Wang and postdoctoral research fellow Andrea Piserchio – all colleagues in the Department of Molecular Pharmacology, Physiology and Biotechnology – focused on the engine. This domain, known as PDZ1, is where the protein links to NMDA receptors. The team used high-resolution nuclear magnetic resonance spectroscopy to solve the structure of PDZ1, as well as a small portion of the receptor to which it binds.
Mierke said the group is now developing a molecule that can inhibit PDZ1 as well as PDZ2, the first boxcar on the multi-domain protein.
The National Institutes of Health funded the work.
Media Contact
More Information:
http://www.brown.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

“Nanostitches” enable lighter and tougher composite materials
In research that may lead to next-generation airplanes and spacecraft, MIT engineers used carbon nanotubes to prevent cracking in multilayered composites. To save on fuel and reduce aircraft emissions, engineers…

Trash to treasure
Researchers turn metal waste into catalyst for hydrogen. Scientists have found a way to transform metal waste into a highly efficient catalyst to make hydrogen from water, a discovery that…
Real-time detection of infectious disease viruses
… by searching for molecular fingerprinting. A research team consisting of Professor Kyoung-Duck Park and Taeyoung Moon and Huitae Joo, PhD candidates, from the Department of Physics at Pohang University…





















