Same fold in viral shells point to common ancestry
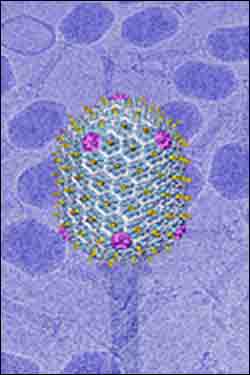
This figure shows the capsid, or outer shell, of a virus called bacteriophage T4. Proteins called gp23 are represented in blue and gp24 are represented in magenta. Both of these proteins are critical for the assembly of the T4 capsid. Researchers have found that the T4 virus and another virus called HK97 both have similar "protein folds" in their outer shells. The findings are providing further evidence that the protein envelope protecting DNA in viruses evolved billions of years ago from a common ancestor and uses the same basic protein fold to construct the outer shell. The researchers used X-ray crystallography to view the gp24 protein at a resolution of 2.9 angstroms and electron microscopy to view the virus capsid at 22-angstrom resolution. An angstrom is one ten-billionth of a meter, or roughly one-millionth as wide as a human hair. (Image courtesy of Purdue University Department of Biological Sciences)
New findings in research led by Purdue University biologists provide further evidence that the protein envelope protecting DNA in viruses evolved billions of years ago from a common ancestor and uses the same basic protein “fold” to construct the critical outer shell.
The most recent findings, which appear in the current issue of the Proceedings of the National Academy of Sciences, show that the T4 virus has a similar protein fold in its outer shell, or capsid, as another virus called HK97.
The protein fold of the viral envelope is crucial for the assembly of the capsid, which protects DNA vital to a virus’ ability to infect host organisms and reproduce. The T4 and HK97 viruses are called bacteriophages because they infect bacteria.
“In the case of the T4 virus, the capsid is made of interconnected six-sided structures called hexamers, which link together in a honeycomb-like pattern to form a very thin wall that is incredibly stable,” said Michael Rossmann, the Hanley Distinguished Professor of Biological Sciences in Purdue’s College of Science. “The capsid surrounds and protects the virus’ DNA, and you can think of it as a balloon that doesn’t burst even though you are pumping in more and more air. “It has now been shown that many viruses have derived their protein envelope from a common ancestor.”
The research was performed by Andrei Fokine and Petr Leiman, postdoctoral researchers in Rossmann’s laboratory; Mikhail M. Schneider, in the laboratory of Vadim V. Mesyanzhinov in the Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry in Russia; Bijan Ahvazi and Karen M. Boeshans, in the laboratory of Alasdair C. Steven at the National Institute of Arthritis and Musculoskeletal and Skin Diseases in Bethesda, Md.; and Lindsay W. Black, from the University of Maryland School of Medicine in Baltimore.
The findings were posted online May 6.
“This work marks only the second time researchers have shown in atomic detail that different bacteriophages have a similar fold,” Rossmann said. “That’s because we and the original discoverers of the special fold in HK97, Jack Johnson and colleagues from the Scripps Research Institute in La Jolla, Calif., used X-ray crystallography, which leaves no doubt about the atomic structure.”
Capsids contain proteins, which are made of a string of building blocks called amino acids. Proteins fold into specific shapes, depending mostly on the sequence of amino acids. “If capsid folds have similar structures and amino acid sequences, then they probably have evolved from a common ancestor,” Rossmann said.
Purdue structural biologists led by Rossmann wrote a paper published last month announcing they had determined that the phi 29 virus, which attacks the soil bacterium bacillus subtilis, possesses a protein fold in its capsid similar to the fold found in the HK97 virus, which infects E. coli bacteria.
Researchers used a technique called cryoelectron microscopy to determine the three-dimensional structure of phi 29 down to a resolution of 7.9 angstroms. An angstrom is one ten-billionth of a meter, or roughly one-millionth as wide as a human hair.
The new paper details how the researchers used X-ray crystallography to analyze the structure of a protein called gp24, which is a key part of the capsid fold in the T4 virus. X-ray crystallography yields more precise results because it is capable of higher resolution than cryoelectron microscopy. The researchers used X-ray crystallography to view the gp24 protein at a resolution of 2.9 angstroms.
The X-ray crystallography data confirmed that gp24 has a fold similar to that of the HK97 capsid protein and also revealed the structure of an “insertion domain,” or a portion of gp24 that may play a role in connecting adjacent subunits to assemble the capsid. Finding the gp24 structure also enabled the researchers to deduce the structure of another protein, gp23, which is crucial along with gp24 in forming the hexamer segments of the capsid.
Researchers are studying the T4 virus for many fundamental biological studies, some of which might be used to create new vaccines to fight viral infections in humans. “Many people have talked about using T4 or other bacteria viruses as antibiotics,” Fokine said. “Although we are conducting research regarding T4’s potential vaccine applications, that is not a focus of the work being reported in this paper.”
The T4 capsid is made up of five proteins, which come together like the pieces of a jigsaw puzzle.
One of the proteins in the T4 capsid is called a highly antigenic outer capsid protein, or hoc. Proteins that mimic proteins from dangerous viruses might be attached to this hoc protein. Then the altered T4 virus could be administered as a vaccine, prompting an immune response against the harmful viruses.
In collaboration with V. Rao of the Catholic University of America in Washington, D.C., Fokine and Rossmann are conducting research aimed at using the T4 capsid as a vaccine for anthrax.
To study the structure of gp24, the researchers first turned the protein into a crystal form and then analyzed it with X-ray crystallography, a technique in which X-rays pass through a crystal, creating a “diffraction pattern” that can be interpreted with various computational procedures.
“Once we determined the structure of gp24, we were able to figure out the shape of gp23 because we know they have a similar structure from the similarity of their amino acid sequences,” Rossmann said.
The researchers needed to know the structures of both gp23 and gp24 before they could learn the overall organization of the T4 capsid. Other scientists, Wah Chiu and his colleagues at Baylor School of Medicine in Houston, had previously found that another bacteriophage, called P22, contains a similar capsid fold as HK97. The Purdue team has since discovered the same fold in phi 29 and the T4 virus, meaning four bacteriophages have been found to possess the fold, bolstering evidence that the fold is common among bacteriophages.
“One fact that makes the T4 research especially interesting is that it is a much larger virus than HK97, P22 and phi29, so the T4 virus is the first large virus to be shown to have a similar fold in its capsid protein, suggesting the fold is likely a common feature in bacteriophages,” Fokine said.
The findings agree with other work in which scientists have discovered that seemingly unrelated viruses that infect mammals use a similar capsid fold.
The research has been funded by the National Science Foundation and the Human Frontier Science Program.
Media Contact
More Information:
http://www.purdue.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Superradiant atoms could push the boundaries of how precisely time can be measured
Superradiant atoms can help us measure time more precisely than ever. In a new study, researchers from the University of Copenhagen present a new method for measuring the time interval,…

Ion thermoelectric conversion devices for near room temperature
The electrode sheet of the thermoelectric device consists of ionic hydrogel, which is sandwiched between the electrodes to form, and the Prussian blue on the electrode undergoes a redox reaction…

Zap Energy achieves 37-million-degree temperatures in a compact device
New publication reports record electron temperatures for a small-scale, sheared-flow-stabilized Z-pinch fusion device. In the nine decades since humans first produced fusion reactions, only a few fusion technologies have demonstrated…





















