A new hypothesis on the origin of ’junk’ DNA

The explosion of “junk” DNA in animals, plants and fungi may be the simple result of their ancestors’ reduced population sizes, according to a new hypothesis proposed by Indiana University Bloomington and University of Oregon scientists in the Nov. 21 issue of Science.
The hypothesis explains a mysterious genetic difference between bacteria and eukaryotes, a giant group of organisms that includes animals, plants, fungi, algae and other protists. Bacteria tend to have extremely lean genomes; their genes barely fit into them, without much genetic material left over. Eukaryotic genomes are a complex mixture of useful genes and useless (“junk”) DNA jammed haphazardly between genes and even within them.
“The evolution of genomic complexity is inevitable,” said IUB biologist Michael Lynch, who led the study. “It’s just that in bacteria, there is a pressure against it — natural selection — which works more efficiently when population sizes are big. Eukaryotes have much smaller population sizes compared to bacteria, and we believe this is the main reason junk DNA sequences are still with us.”
Junk DNA dominates eukaryotic chromosomes. The chromosomal space taken up by just 30 human genes and the DNA within and between those genes could easily accommodate whole bacterial genomes containing 3,000 or 4,000 genes, Lynch said. While some of what geneticists have called junk DNA is turning out to be not so junky after all, Lynch said a substantial fraction of such genetic material probably deserves the name.
Genetic mutations occur in all organisms. But since large-scale mutations — such as the random insertion of large DNA sequences within or between genes — are almost always bad for an organism, Lynch and University of Oregon computer scientist John Conery suggest the only way junk DNA can survive the streamlining force of natural selection is if natural selection’s potency is weakened.
When populations get small, Lynch explained, natural selection becomes less efficient, which makes it possible for extraneous genetic sequences to creep into populations by mutation and stay there. In larger populations, disadvantageous mutations vanish quickly.
Most experts believe that the first eukaryotes, which were probably single-celled, appeared on Earth about 2.5 billion years ago. Multicellular eukaryotes are generally believed to have evolved about 700 million years ago. If Lynch’s and Conery’s explanation of why bacterial and eukaryotic genomes are so different is true, it provides new insights into the genomic characteristics of Earth’s first single-celled and multicellular eukaryotes.
A general rule in nature is that the bigger the species, the less populous it is. With a few exceptions, eukaryotic cells are so big that they make most bacteria look like barnacles on the side of a dinghy. If the first eukaryotes were larger than their bacterial ancestors, as Lynch believes, then their population sizes probably went down. This decrease in eukaryote population sizes is why a burgeoning of large-scale mutations survived natural selection in the first single-celled and multicellular eukaryotes, according to Lynch and Conery.
To estimate long-term population sizes of 50 or so species for which extensive genomic data was available, Lynch and Conery examined “silent-site” mutations. Silent-site mutations are single nucleotide changes within genes that don’t affect the gene product, which is a protein. Because of their unique characteristics, silent-site mutations can’t be significantly influenced by natural selection. The researchers were able to calculate rough estimates of the species’ long-term population sizes by assessing variation in the species’ silent-site nucleotides.
Of the original group of sampled organisms, Lynch and Conery selected a subset of about 30 and calculated, for each organism, the number of genes per total genome size as well as the longevity of gene duplications per total genome size. They also calculated the approximate amount of each organism’s genome taken up by DNA sequences that do not contain genes.
The researchers found that a consistent pattern emerged when genomic characteristics of bacteria and various eukaryotes were plotted against the species’ total genome sizes. Bigger species, such as salmon, humans and mice, tended to have small, long-term population sizes, more genes, more junk DNA and longer-lived gene duplications. Almost without exception, the species found to have large, long-term population sizes, fewer genes, less junk DNA and shorter-lived gene duplications were bacteria.
The data suggest it is genetic drift (an evolutionary force whose main component is randomness), not natural selection, that preserves junk DNA and other extraneous genetic sequences in organisms. When population sizes are large, drift is usually overpowered by natural selection, but when population sizes are small, drift may actually supersede natural selection as the dominant evolutionary force, making it possible for weakly disadvantageous DNA sequences to accumulate.
“As more organisms’ genomes are sequenced, we will continue to look at whether our model is upheld,” Lynch said.
Lynch’s and Conery’s ongoing research on the origins of genomic complexity receives funding from the National Science Foundation and the National Institutes of Health.
To speak with Lynch, contact David Bricker at 812-856-9035 or brickerd@indiana.edu.
“The Origins of Genome Complexity,” Science, Vol. 302, no. 5649
Media Contact
More Information:
http://newsinfo.iu.edu/All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
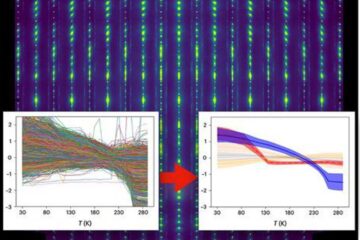
Machine learning algorithm reveals long-theorized glass phase in crystal
Scientists have found evidence of an elusive, glassy phase of matter that emerges when a crystal’s perfect internal pattern is disrupted. X-ray technology and machine learning converge to shed light…
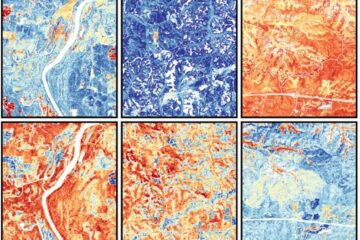
Mapping plant functional diversity from space
HKU ecologists revolutionize ecosystem monitoring with novel field-satellite integration. An international team of researchers, led by Professor Jin WU from the School of Biological Sciences at The University of Hong…

Inverters with constant full load capability
…enable an increase in the performance of electric drives. Overheating components significantly limit the performance of drivetrains in electric vehicles. Inverters in particular are subject to a high thermal load,…





















