Scientists determine identity, cell locale and quantity of nearly all proteins in an organism
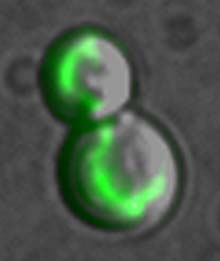
A single protein glows in the mictochondrion of a dividing cell. The mitochondrion is the site of energy production in all higher cells. Only one protein was studied in each of thousands of yeast strains, and the proteins were made to glow florescent green -- a technique that allowed UCSF scientists to identify and pinpoint the location of nearly all the proteins in yeast. They found that more than 500 proteins -- about one in eight in the organism -- are active in the mitochondrion.
First comprehensive view of protein activity in higher organism
UCSF scientists have developed a set of powerful tools that allow researchers to look in unprecedented detail at the full complement of thousands of proteins acting and interacting in a living organism. They have used the new tools to mine nearly the entire proteome of an organism – discovering what proteins are active in each cell, where they are active and in what quantity.
The results, published in two papers in the October 16 issue of the journal Nature, provide the most comprehensive and detailed picture yet of protein activity in the living cells of higher organisms. They reveal a tremendous range in the amount of different proteins active in a cell and help explain the “logic” of the timing and regulation of gene activity.
Recognizing that it is the gene’s products — the proteins – that interact to regulate all life processes, many researchers have begun to turn their attention from the roster of all the genes in an organism – the genome — to the proteome. But while new technologies such as gene chips and whole genome sequencing have accelerated studies of the genome, the technology to study the whole proteome has lagged far behind.
Unlike genes, which all use the same code, each protein has unique physical properties, thwarting attempts to develop a single method to study them all in a living cell. By giving each protein a common tag, the UCSF scientists were able to follow where each protein acted and in what quantity.
With this new method for a thorough protein search, they were able to examine thousands of proteins individually — one in each of thousands of different strains of the common baker’s yeast, Saccharomyces cerevisiae, an organism that has proven to be a good model for understanding basic biology of human cells. About one third of the yeast’s genes are shared by humans, and each yeast cell contains a nucleus and other basic organelles that function in humans cells.
By scrutinizing only one protein in each strain under study, the researchers determined precisely where and in what quantity each protein acts during normal growth — a tool that can now be used to identify the role of different proteins, the relationship between sets of genes and proteins, and the links between protein changes and disease.
The new approach was far more sensitive than any previous attempt to detect low levels of protein activity, the scientists reported. They detected proteins in quantities as low as 50 molecules per cell – an extraordinarily small amount to detect — and as high as a million or more molecules per cell, demonstrating that living cells utilize different proteins in vastly different amounts.
By integrating the new protein findings with those of gene studies, the scientists were able to decipher some of the logic by which certain genes are normally turned on and off together. They found that such “co-regulated” genes code for proteins that reside in the same place in the cell.
“Things make more sense when you can look at the level of proteins – the actual players in the life of cells and organisms,” said Jonathan Weissman, PhD, a Howard Hughes Medical Institute (HHMI) investigator, professor of cellular and molecular pharmacology at UCSF, and co-senior author on both papers.
The gene messages are “imperfect proxies” for the information we really care about, which is the proteins,” Weissman added. “The reason we’ve been looking at the messages – the genes — is because we could, but what is more informative is proteins – where they act, how they act and in what kind of abundance.”
“There has been a real lack of good tools to follow what proteins were doing in a cell in any comprehensive way. Because of this, fundamental questions remained unanswerable about the proteome and its relationship to the genome. Now, we can address those questions.”
Altogether, 4,200 proteins were identified -about 80 percent of all the different proteins in the baker’s yeast.
Erin O’Shea, PhD, also an HHMI investigator and UCSF professor of biochemistry and biophysics, is also co-senior author on both papers.
The scientists used two identical sets of thousands of strains of the yeast — one to determine where in the cell each protein acts, and the other to determine the abundance of each protein. In the study of each protein’s cellular locale, they “tagged” a different gene in each yeast strain.
The tag was a green florescent marker, attached to the gene in such a way that it was not likely to disrupt the gene’s function. When the gene carried out its normal role, instructing the cell to make a specific protein, that protein glowed green, and could be located in the living cell using microscopy. Since each yeast strain had only one tagged protein, the researchers could pinpoint the location of nearly all of the organism’s proteins by looking for the telltale green sign in each strain.
At the outset, they assumed that yeast contain about 6,200 proteins, since other scientists had identified that many genes in DNA, but through this and a second tagging approach, the researchers found that only about 5,500 of the suspected genes actually coded for proteins.
They used the second collection of yeast strains to tag the proteins with a standard marker that allows purification and measurement of the abundance of the protein. When the results of the two parallel approaches were pooled, the team was able to identify the type, cell location and abundance of about 80 percent of the yeast’s proteins.
The scientists were not able to study all of the intended 5,500 proteins. The study could only identify a protein if its genes expressed it – if the protein was active. Yeast have at least two major phases in their life cycle: the main period of feeding, growth and reproduction, and at least one phase in which the organism responds to specialized stress conditions and produces spores – sort of like abandoning ship. The scientists expect that a significant proportion of the proteins they did not identify simply are not expressed in the normal growth phase that they studied.
In addition, some proteins may be expressed in such small amounts – less than 50 molecules per cell, for example – that they are below the detection level of even this highly sensitive approach. Finally, for strictly technical reasons, some of the proteins may have escaped successful tagging.
Nonetheless, the project was easily thorough enough to net some striking findings. For example, the research revealed that of the 4,200 yeast proteins scrutinized, a full 527 of them — or about one in eight – work in the energy-converting organelle, the mitochondrion. O’Shea speculates that the mitochondrion may have such a large share of the proteins because, in addition to it vital energy conversion function, this organelle is believed to be derived from a separate organism – a bacterial parasite or pathogen that invaded our ancient ancestral cell. The mitochondrion, as scientists now believe, is a direct descendent of an independent organism that simply “moved in” and started keeping house with its host.
Weissman and O’Shea have known each other since graduate school in 1989. Both were named Howard Hughes Medical Institute investigators at UCSF in 2000 -prestigious and generous support for biomedical scientists.
“HHMI urged all the recipients to do something particularly challenging,” O’Shea says. “So, Jonathan and I decided to pool our HHMI resources to try to develop the tools need to study proteomics.”
“Genomics was led by technology that we didn’t have before,” Weissman says. “Proteomics can’t advance unless we develop new technology to study thousands of proteins in a systematic way. That’s what we set out to do.”
The research was completed at the Center for Advanced Technology at UCSF’s new Mission Bay campus. The CAT is intended to support development of new technologies.
Lead authors on the Nature article, “Global Analysis of Protein Localization in Budding Yeast” are Won-Ki Huh, PhD and James Falvo, PhD, postdoctoral fellows in O’Shea’s lab. Co-authors are Luke C. Gerke, BS, research associate; Adam S. Carroll, PhD, research specialist; and Russell W. Howson, graduate student.
Lead author on the Nature letter in the same issue, “Global Analysis of Protein Expression in Budding Yeast,” is Sina Ghaemmaghami, PhD, postdoctoral fellow in Weissman’s lab. Co-authors are Won-Ki Huh; Kiowa Bower, BA, research technician; Russell Howson; Archana Belle, PhD, postdoctoral fellow; and graduate student Noah Dephoure.
In addition to the Howard Hughes Medical Institute, the research was supported by the David and Lucille Packard Foundation.
Media Contact
More Information:
http://www.ucsf.edu/All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Silicon Carbide Innovation Alliance to drive industrial-scale semiconductor work
Known for its ability to withstand extreme environments and high voltages, silicon carbide (SiC) is a semiconducting material made up of silicon and carbon atoms arranged into crystals that is…

New SPECT/CT technique shows impressive biomarker identification
…offers increased access for prostate cancer patients. A novel SPECT/CT acquisition method can accurately detect radiopharmaceutical biodistribution in a convenient manner for prostate cancer patients, opening the door for more…

How 3D printers can give robots a soft touch
Soft skin coverings and touch sensors have emerged as a promising feature for robots that are both safer and more intuitive for human interaction, but they are expensive and difficult…





















