Purdue team solves structure of West Nile virus
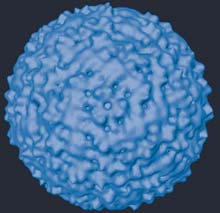
This figure shows a surface-shaded image of the West Nile virus particle produced by Purdue University biologists using cryoelectron microscopy. The surface is composed of proteins that enable the virus to bind with and invade a host cell. The particle is approximately 50 nanometers in diameter, or about 1/1000th of the width of a human hair. (Purdue Department of Biological Sciences image) <br>
Purdue University biologists have determined the structure of the West Nile virus, a development that could greatly augment our understanding of the virus’ life cycle.
Using cryoelectron microscopy and advanced imaging techniques, the Purdue team has determined the orientation of the major surface proteins in a West Nile viral particle. Because these proteins are instrumental in allowing the virus to bind to and invade a host cell, the research could be a step forward in combating the deadly mosquito-borne disease.
“We can now clearly understand how these proteins interact with one another,” said Richard J. Kuhn, a professor of biological sciences in Purdue’s School of Science. “We can’t cure West Nile yet, but we can now start thinking about how to interfere with these interactions, which could be a key to stopping the infection’s progress.”
The team’s work appears in Friday’s (10/10) edition of Science.
Viruses are among the smallest of biological entities, containing only essential amounts of genetic material that allow a virus to take over a victim cell’s functions. As West Nile develops inside a host cell, several layers of protein molecules assemble themselves around the genetic material, forming a protective shell. The outer layer of proteins is often arranged in an intricate pattern of interlocked molecules that can give the particle’s surface the appearance of a lattice or, in the case of West Nile, the fabric of a herringbone jacket. When the mature West Nile virus particle emerges, it is these surface proteins that interact with another cell’s surface so the next invasion cycle can begin.
“The West Nile virus is formed from three protein types,” Kuhn said. “After the virus assembles in its host cell, these protein molecules fit together like a jigsaw puzzle and form a well-ordered symmetrical particle. From the structure, we now know, essentially, how the major sets of protein molecules interlock with each other chemically. Armed with this knowledge, scientists might now conceive of ways to interrupt the viral assembly process.”
Adding to the knowledge base is the previous work the group has done with flaviviruses, the viral family that causes diseases including West Nile, dengue and yellow fever. The group, which is composed of researchers from Kuhn’s lab, as well as the labs of Michael Rossmann and Timothy Baker, has described the structure of other flaviviruses before (see related articles below). While this is the first time West Nile’s structure has been described, Kuhn said the group’s past work could assist with science’s understanding of this particular viral family.
“What we already know from studying other flaviviruses could give us a leg up understanding West Nile’s behavior,” he said. “Dengue, for example, has a very similar structure to West Nile’s, but its surface features are sufficiently different that comparisons could help shed light on how West Nile operates.”
Since it first appeared in New York City in 1999, West Nile virus has spread to 44 states, hitting the northern plains and eastern Rocky Mountains particularly hard. So far this year, there have been more than 4,400 cases and 84 deaths nationwide. The Centers for Disease Control and Prevention maintains a Web site with information on the spread of West Nile and its prevention.
While Kuhn is hopeful that the group’s work will add to the effort to contain the disease, he said much additional work will be required to understand the virus’ life cycle on the molecular level.
“Our structural map now shows only the general orientation of the proteins,” he said. “What we need now is to include what you might think of as an ’inset map’ – an even smaller-scale picture that details the structure of each of the three protein varieties that make up the virus particle. Then researchers will have more insight into how the proteins bond with cells and each other.”
This closer look represents the next step for Purdue’s structural virology group, which has recently received new support from the National Institutes of Health in the form of grants totaling nearly $18 million. These grants support basic research on viral infectious diseases, including West Nile.
“We currently have a 17-angstrom resolution structure,” Kuhn said. “We hope to use our NIH support to get down to the 9-angstrom scale or better, as this would give us details on the individual protein molecules as well as other proteins in the virus.”
The work on the West Nile virus was performed by Suchetana Mukhopadhyay, Bong-Suk Kim and Paul R. Chipman, who are associated with the labs of Rossmann and Kuhn. The group is part of the Markey Center for Structural Biology, which consists of laboratories that use a combination of cryoelectron microscopy, crystallography, nuclear magnetic resonance, biochemistry and molecular biology to elucidate the processes of viral entry, replication and pathogenesis. Approximately 100 research scientists, including 50 graduate students, comprise the Markey Center.
This work was funded by the National Institute of Allergy and Infectious Diseases.
Writer: Chad Boutin, (765) 494-2081, cboutin@purdue.edu
Source: Richard J. Kuhn, (765) 494-1164, kuhnr@purdue.edu
Purdue News Service: (765) 494-2096; purduenews@purdue.edu
Media Contact
More Information:
http://news.uns.purdue.edu/html4ever/031009.Kuhn.westnile.htmlAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Optimising inventory management
Crateflow enables accurate AI-based demand forecasts. A key challenge for companies is to control overstock and understock while developing a supply chain that is resilient to disruptions. To address this,…

Cause of rare congenital lung malformations
Gene mutations in the RAS-MAPK signaling pathway disrupt lung development in the womb. Most rare diseases are congenital – including CPAM (congenital pulmonary airway malformations). These are airway malformations of…

“Nanostitches” enable lighter and tougher composite materials
In research that may lead to next-generation airplanes and spacecraft, MIT engineers used carbon nanotubes to prevent cracking in multilayered composites. To save on fuel and reduce aircraft emissions, engineers…





















