New "Knockout" Map Helps Study Gene Functions in Model Plant

The image displays a single Arabidopsis mutant line from the Salk Institute insertion mutant collection/database. The location of the Agrobactrium T-DNA insertion is known from sequencing of The image displays a single Arabidopsis mutant line from the Salk Institute insertion mutant collection/database. The location of the Agrobactrium T-DNA insertion is known from sequencing of the genome. <br>Credit: Kent Schnoeker, The Salk Institute
Scientists have inactivated almost three-quarters of all genes in the genome of Arabidopsis thaliana, a species widely used in plant research. The feat, which results in the largest so-called “knockout” gene collection of a complex multi-cellular organism, now allows researchers to study the function of each of those genes individually or together.
The findings, published in the August 1 issue of the journal Science, mark an important milestone in the field of plant genomics. Following the release of the Arabidopsis genome sequence in 2000, the National Science Foundation (NSF) jump started the next phase of plant genome research, instituting the Arabidopsis 2010 Project to determine the location and function of each and every Arabidopsis gene by the year 2010. Knowing the function of all the genes in this model plant will aide scientists immensely in their work to improve disease resistance, control how quickly or slowly fruit will ripen, and create healthier and improved crops.
Joe Ecker at the Salk Institute for Biological Studies and his research team created knockouts, or inactivating mutations, in 21,700 of the estimated 29,454 Arabidopsis genes. Knocking out a gene or group of genes allows scientists to observe what goes wrong in the mutant plant and determine what function the inactivated gene(s) had in the plant system.
“This project provides a resource that is essential for understanding the function of genes – mutants in which individual genes have been inactivated,” says Parag Chitnis, program officer in the NSF’s Division of Molecular and Cellular Biosciences. “Because the expression of hundreds of genes can change in a single condition…, large complete collections of stable mutations are becoming an important tool for whole genome function and evolutionary biology,” the report further emphasizes.
History was made in December of 2000 when the Arabidopsis research community released the complete sequence of the mustard family weed. Yet the exact location and function of the nearly 29,500 Arabidopsis genes remained unknown. Using the plant’s genome sequence in conjunction with new technologies in the field, the Salk team identified the precise locations of more than 88,000 mutations they made in the Arabidopsis genome. That advance nearly completes a key step in the 2010 project. But before the function of all Arabidopsis genes can be determined, a knockout mutant plant must be created for every single gene in the plant’s genome.
To create a gene knockout, scientists use a bacterium called Agrobacterium to insert a code that tells a specific gene to turn off. According to Ecker, this process of T-DNA integration has been carried out for well over 25 years, but this study provides a new perspective on using the technique to analyze gene function.
Some genes, it turns out, contain certain features that mark them as favored targets of inactivation. Additionally, Ecker and his colleagues have discovered that fewer inactivations occur near the centromeres — the thinner gene-poor regions of the chromosome. “These results provide significant new information in both the areas of functional genomics and basic plant biology,” says Ecker.
Because Arabidopsis is a model plant, knowledge gained from this plant can be applied to many other plants as well. Ecker is optimistic about the progress of this project. “The information you get from Arabidopsis is very likely to be immediately applicable to all plants. The information that we produce will be used by a range of people to improve plant growth, yield, and drought tolerance.”
The database of Arabidopsis gene knockouts completed thus far is available to the public, as are all corresponding strains of mutant plants created by the research team, which includes scientists from the Salk Institute, the NRC Plant Biotechnology Institute, and UC San Diego. Funding was provided by the National Science Foundation Arabidopsis 2010 Project and the Department of Energy.
Principal Investigator: Joseph Ecker, +1-858-453-4100 ext.1752, ecker@salk.edu
Media Contact
More Information:
http://signal.salk.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
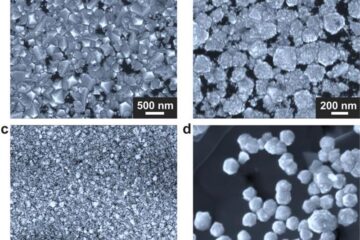
Making diamonds at ambient pressure
Scientists develop novel liquid metal alloy system to synthesize diamond under moderate conditions. Did you know that 99% of synthetic diamonds are currently produced using high-pressure and high-temperature (HPHT) methods?[2]…

Eruption of mega-magnetic star lights up nearby galaxy
Thanks to ESA satellites, an international team including UNIGE researchers has detected a giant eruption coming from a magnetar, an extremely magnetic neutron star. While ESA’s satellite INTEGRAL was observing…
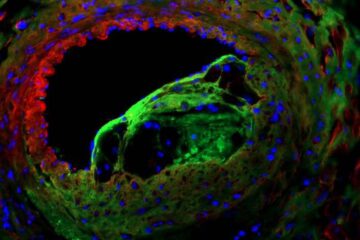
Solving the riddle of the sphingolipids in coronary artery disease
Weill Cornell Medicine investigators have uncovered a way to unleash in blood vessels the protective effects of a type of fat-related molecule known as a sphingolipid, suggesting a promising new…





















