Researchers Are First To Simulate The Binding Of Molecules To A Protein

Now researchers at the University of Illinois have identified a key step in the cellular recycling of ATP that allows your body to produce enough of it to survive. Without this cycling of ATP and its low-energy counterpart, adenosine diphosphate (ADP), into and out of the mitochondrion, where ADP is converted into ATP, life as we know it would end.
Researchers have for the first time simulated the binding of ADP to a carrier protein lodged in the inner membrane of the mitochondrion. It is the first simulation of the binding of a molecule to a protein. Their findings appear this week in Proceedings of the National Academy of Sciences.
As its name indicates, ATP contains three phosphate groups. The energy produced when one of these groups is detached from the molecule drives many chemical reactions in the cell. This process also yields ADP, which must go through the ADP/ATP carrier (AAC) to get into the mitochondrion to be converted back into ATP.
The AAC acts a lot like a revolving door: For each molecule of ADP going into the mitochondrion, one ATP gets booted out. These two activities are not simultaneous, however. The carrier is either shuttling ADP into the mitochondrion or ejecting ATP into the wider environment of the cell, where it can be put to use.
“The carrier is a reversible machine,” said biochemistry professor Emad Tajkhorshid, who led the study which was conducted by biophysics graduate student Yi Wang. “Both ATP and ADP can bind to it and make it to the other side using this transporter.”
Previous studies used X-ray crystallography to determine the three-dimensional structure of the carrier when it was ready to accept a molecule of ADP.
In the new analysis, the researchers developed a computer simulation of the interaction of a single molecule of ADP with the carrier protein. Thanks to better simulation software and larger and more sophisticated computer arrays than were available for previous studies, this simulation tracked the process by which ADP is drawn into the carrier. It also showed how ADP orients itself as it travels to the site where it binds to the carrier.
In the simulation, the researchers observed for the first time that ADP disrupts several ionic bonds, called salt bridges, when it binds to the carrier protein. Breaking the salt bridges allows the protein to open – in effect unlocking the door that otherwise blocks ADP’s route into the mitochondrion.
The simulation included every atom of the carrier protein and ADP, as well as all of the membrane lipids and water molecules that make up their immediate environment – more than 100,000 atoms in all. It tracked the interaction over a period of 0.1 microseconds, an order of magnitude longer than what had been possible before.
“Until two years ago 10 nanoseconds was really pushing it,” Tajkhorshid said. “Now we are reaching the sub-microsecond regime, and that’s why we are seeing more biologically relevant events in our simulations.”
The longer time frame meant that the researchers did not need to manipulate the interaction between the molecules. They simply positioned the ADP at the mouth of the carrier protein, some 25 angstroms from the site where they knew it was meant to bind. (An angstrom is one ten-millionth of a meter. Most molecular binding interactions occur at less than 6 or 7 angstroms.) They even placed the ADP upside-down at the mouth of the protein carrier and saw it flip into an orientation that allowed it to bind to the carrier.
The identified binding pocket for ADP explained a lot of known experimental data, and revealed an unusual feature of the carrier protein: Its binding site and the entryway leading to it had an extremely positive electrical charge.
It had a much greater positive charge than any known protein transporter.
This positive charge appears to serve two functions, Tajkhorshid said. First, it allows the protein carrier itself to nestle tightly in the mitochondrial membrane, which contains a lot of negatively charged lipids. Second, it strongly attracts ADP, which carries a negative charge. More interestingly, through a bioinformatics analysis the researchers show that this unusual electrostatic feature is common to all mitochondrial carriers.
Other negatively charged ions can enter the carrier, Tajkhorshid said, but only a molecule with at least two phosphate groups can disrupt the salt bridges to activate it.
This simulation marks the first time that researchers have been able to describe in molecular detail how a protein binds to the molecule that activates it, Tajkhorshid said.
The findings shed light on a fundamental physiological process, he said.
“Any time you move anything in your body, you use ATP,” he said. “Many enzymatic reactions also require ATP. In the central nervous system, the transport of hormones, neurotransmitters or other molecules, these are all ATP-dependent.”
“It has been estimated that you burn more than your body weight in ATP every day,” he said. “So that’s how much ATP you have to carry across the inner mitochondrial membrane every day – through this guy.”
Tajkhorshid is also a professor of pharmacology in the College of Medicine and an affiliate of the Beckman Institute and the Center for Biophysics and Computational Biology in the College of Liberal Arts and Sciences.
Media Contact
More Information:
http://www.uiuc.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
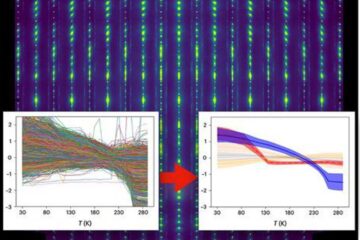
Machine learning algorithm reveals long-theorized glass phase in crystal
Scientists have found evidence of an elusive, glassy phase of matter that emerges when a crystal’s perfect internal pattern is disrupted. X-ray technology and machine learning converge to shed light…
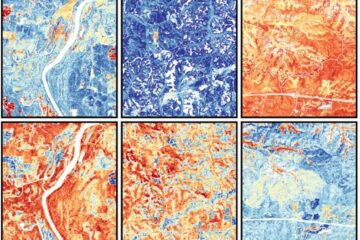
Mapping plant functional diversity from space
HKU ecologists revolutionize ecosystem monitoring with novel field-satellite integration. An international team of researchers, led by Professor Jin WU from the School of Biological Sciences at The University of Hong…

Inverters with constant full load capability
…enable an increase in the performance of electric drives. Overheating components significantly limit the performance of drivetrains in electric vehicles. Inverters in particular are subject to a high thermal load,…





















