Protein could put antibiotic-resistant bugs in handcuffs
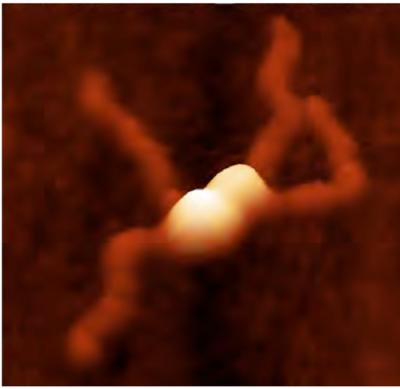
An atomic force microscopy image shows a four-part complex of the protein RepA (bright mounds) bound together and "handcuffing" two thread-like strands of DNA plasmid. Credit: University of Nebraska Medical Center Nanoimaging Core Facility
Staph infections that become resistant to multiple antibiotics don't happen because the bacteria themselves adapt to the drugs, but because of a kind of genetic parasite they carry called a plasmid that helps its host survive the antibiotics.
Plasmids are rings of bare DNA containing a handful of genes that are essentially freeloaders, borrowing most of what they need to live from their bacterial host. The plasmids copy themselves and go along for the ride when the bacteria divide to copy themselves.
A team from Duke and the University of Sydney in Australia has solved the structure of a key protein that drives DNA copying in the plasmids that make staphylococcus bacteria antibiotic-resistant. Knowing how this protein works may now help researchers devise new ways to stop the plasmids from spreading antibiotic resistance in staph by preventing the plasmids from copying themselves.
“If plasmids can't replicate, they go away,” said lead author Maria Schumacher, an associate professor of biochemistry in the Duke University School of Medicine. “This is a fantastic new target for antibiotics.”
The work appears the week of June 9 in the Proceedings of the National Academy of Sciences.
An essential part of biology, plasmids are so minimalistic they're not even considered alive by themselves. But they're good at ferrying genes from one kind of bacteria to another in a process called horizontal gene transfer. They also excel at adapting to environmental conditions more quickly than their bacterial hosts. Plasmids are able to develop new defenses to an antibiotic and then share that new trick with other bacteria.
Through several years of laborious structural biology to figure out the specific shapes of the molecules involved, the research team has mapped out the structure and function of a protein called RepA, which is crucial to the plasmids' ability to copy its DNA and make a new plasmid.
RepA is a protein that sticks to the beginning of the plasmid's DNA sequence and starts the copying process. “This protein is essential to everything,” Schumacher said. “If you don't have it, the plasmid will quickly cease to exist.”
Plasmids also need a mechanism to prevent themselves from making too many copies, which would strangle their bacterial host. The researchers have found that RepA is crucial to that function as well.
RepA naturally sticks together in pairs. When a pair of RepA proteins bumps into another pair, as when the cell is starting to get crowded with plasmids, the two pairs of RepA preferentially stick to each other. They form a complex back-to-back, with both having their DNA-grabbing parts facing outward.
When RepA forms this four-part molecule, the plasmids are said to be 'handcuffed,' because two rings of DNA are captured with the locked-up and non-functional RepA complex in the middle.
Once it is handcuffed like this, the plasmid will no longer replicate. Schumacher said this mechanism is apparently how RepA prevents the plasmids from overpopulating the bacterial cell.
Schumacher says RepA is ubiquitous in the plasmid world and doesn't bear much resemblance to other proteins, or to human proteins, making it an attractive drug target. She is hopeful the molecule could be a new site to attack with antibiotics.
“This has been a fun project because we saw many things we didn't expect to see,” Schumacher said.
The research was supported by the National Institutes of Health and Department of Energy in the U.S., and the National Health and Medical Research Council of Australia.
CITATION: “Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein,” Maria Schumacher, Nam K. Tonthat, Stephen M. Kwon, Nagababu Chinnam, Michael A. Liu, Ronald A. Skurray and Neville Firth. Proceedings of the National Academy of Sciences, June 9, 2014. DOI: 10.1073/pnas.1406065111
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

High-energy-density aqueous battery based on halogen multi-electron transfer
Traditional non-aqueous lithium-ion batteries have a high energy density, but their safety is compromised due to the flammable organic electrolytes they utilize. Aqueous batteries use water as the solvent for…

First-ever combined heart pump and pig kidney transplant
…gives new hope to patient with terminal illness. Surgeons at NYU Langone Health performed the first-ever combined mechanical heart pump and gene-edited pig kidney transplant surgery in a 54-year-old woman…

Biophysics: Testing how well biomarkers work
LMU researchers have developed a method to determine how reliably target proteins can be labeled using super-resolution fluorescence microscopy. Modern microscopy techniques make it possible to examine the inner workings…





















