Mechanism to Repair Clumped Proteins Explained
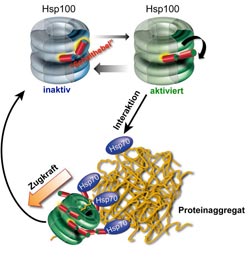
Mechanism of protein aggregate dissolution through Hsp70/Hsp100 cooperation. The ring-shaped Hsp100 has two structural states, one inactive and the other activated. A molecular switch keeps the Hsp100 chaperone in the inactive state. Hsp70 causes the switch to flip, thereby activating the Hsp100 chaperone. In this state it can pull protein strands out of the aggregate. The activation of Hsp100 is not permanent, with the chaperone reverting to the inactive state after the aggregate has dissolved.<br><br>Picture credits: ZMBH<br>
Clumped proteins can be dissolved with the aid of cellular repair systems – a process of critical importance for cell survival especially under conditions of stress. Heidelberg researchers have now decrypted the fundamental mechanism for dissolving protein aggregates that involves specific molecular chaperones.
Scientists from the Center for Molecular Biology of Heidelberg University and the German Cancer Research Center cooperated with experts from the Heidelberg Institute for Theoretical Studies on the project. The results of the research appeared in two simultaneously published articles in “Nature Structural & Molecular Biology”.
Proteins consist of long chains of successive amino acids and perform vital functions in every cell. To function, every amino acid chain must first assume a specific three-dimensional structure – it has to fold itself. A change in growth conditions, such as an increase in ambient temperature, can cause proteins to lose their structure and unfold. Unfolded protein chains run the risk of clumping, forming protein aggregates. “If such aggregates form, the proteins cannot function, which can lead to cell death, which we see in neurodegenerative diseases such as Alzheimer’s and Parkinson’s, and even in ageing processes”, explains Prof. Dr. Bernd Bukau, Director of the Center for Molecular Biology of Heidelberg University (ZMBH), who is also a researcher at the German Cancer Research Center (DKFZ).
But clumping does not necessarily mean the end of a protein’s life cycle. “Cells have repair systems for damaged proteins, so-called molecular chaperones, that can dissolve even aggregated proteins and refold them”, clarifies Dr. Axel Mogk, also a member of the ZMBH and DKFZ. The repair is carried out by a cooperating team of two chaperones, called Hsp70 and Hsp100. The Heidelberg researchers were able to demonstrate that the activity of the Hsp100 chaperone is regulated by a built-in molecular switch.
This switch is first positioned to curtail energy consumption, i.e. ATP hydrolysis, and thereby the activity of the Hsp100 chaperone. The cooperating Hsp70 protein changes the position of the switch and activates Hsp100 directly at the protein aggregate. In this state, the “motor” of the ring-shaped Hsp100 protein runs at full speed, reaches top performance and is able to extract individual chains from the aggregate. Afterwards, the extracted, unfolded protein can start the folding process over. The results of the Heidelberg research also show that the built-in switch’s control of Hsp100 activity is of vital importance for this complicated protein machine, because the loss of regulation in hyperactive, i.e. permanently activated, Hsp100 protein variants leads to cell death.
The research collaboration falls under the DKFZ-ZMBH Alliance, the strategic cooperation of the German Cancer Research Center and the Center for Molecular Biology of Heidelberg University. The Heidelberg Institute for Theoretical Studies (HITS) develops new theoretical approaches to interpreting the burgeoning amount of experimental data.
Original publications:
F. Seyffer, E. Kummer, Y. Oguchi, J. Winkler, M. Kumar, R. Zahn, V. Sourjik, B. Bukau & A. Mogk: Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces, Nature Structural & Molecular Biology, 18 November 2012, doi: 10.1038/nsmb.2442
Y. Oguchi, E. Kummer, F. Seyffer, M. Berynskyy, B. Anstett, R. Zahn, R.C. Wade, A. Mogk & B. Bukau: A tightly regulated molecular toggle controls AAA+ disaggregase, Nature Structural & Molecular Biology, 18 November 2012, doi: 10.1038/nsmb.2441
Contact:
Prof. Dr. Bernd Bukau, Dr. Axel Mogk
Center for Molecular Biology of Heidelberg University
Phone: +49 (0)6221 54-6850, direktor@zmbh.uni-heidelberg.de
Phone: +49 (0)6221 54-6863, a.mogk@zmbh.uni-heidelberg.de
Communications and Marketing
Press Office, phone: +49 (0)6221 54-2311
presse@rektorat.uni-heidelberg.de
Media Contact
More Information:
http://www.uni-heidelberg.deAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Superradiant atoms could push the boundaries of how precisely time can be measured
Superradiant atoms can help us measure time more precisely than ever. In a new study, researchers from the University of Copenhagen present a new method for measuring the time interval,…

Ion thermoelectric conversion devices for near room temperature
The electrode sheet of the thermoelectric device consists of ionic hydrogel, which is sandwiched between the electrodes to form, and the Prussian blue on the electrode undergoes a redox reaction…

Zap Energy achieves 37-million-degree temperatures in a compact device
New publication reports record electron temperatures for a small-scale, sheared-flow-stabilized Z-pinch fusion device. In the nine decades since humans first produced fusion reactions, only a few fusion technologies have demonstrated…





















