How Fic proteins regulate their potentially lethal enzyme activity
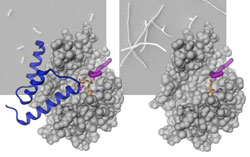
Left: Binding of the antitoxin (blue) inhibits AMPylation of the target protein (magenta) by the Fic protein (grey), which allows normal bacterial growth. Right: In the absence of the antitoxin the target protein gets AMPylated, resulting in inhibition of cell division and thus abnormal filamentous growth of bacteria. Illustration: Universität Basel<br>
The research groups headed by Prof. Christoph Dehio and Prof. Tilman Schirmer could demonstrate that through the alteration of one single amino acid this inhibition of enzyme activity can be relieved. Their findings, which have been published in the current issue of «Nature», will enable to investigate the physiological role of the potentially lethal function of Fic proteins in bacteria and higher organisms in the future.
Fic proteins are found in most forms of life ranging from simple bacteria to man. Only a few representatives of this protein family of about 3000 members have been investigated to date. These are enzymes that chemically alter other proteins through the attachment of an adenosine monophosphate group (AMP) derived from the important energy carrier ATP. This reaction, known as AMPylation, specifically modifies the function of the target proteins.
The biochemically best understood Fic proteins are produced by pathogenic bacteria and injected into host cells to alter cellular signaling proteins to the advantage of the bacterial intruder. However, the far majority of Fic proteins have probably evolved a function that is instrumental for the cell in which they are produced. Why the biochemical function of only a few of these Fic proteins has been elucidated so far was not clear. The reason has now been found by the collaborating research groups of the infection biologist Prof. Christoph Dehio and the structural biologist Prof. Tilman Schirmer.
The Active Center of Fic Proteins is Blocked
The scientists could show that an amino acid residue (glutamate-finger) protrudes into the active center of the Fic proteins. This prevents productive binding of ATP and explains the inactivate ground state of the enzyme. Surprisingly, in some Fic proteins the inhibiting residue is part of the Fic protein itself, whereas in other cases it is provided by a separate protein (called antitoxin). It was shown that upon truncation of the glutamate-finger by genetic manipulation or removal of the entire antitoxin the activity of the enzyme is awakened – sometimes with drastic consequences for the affected cells. Bacterial cells no longer divide, while human cells can even die.
Interdisciplinary Research Success
The two research groups achieved this breakthrough by combining methods of microbiology, cell biology, structural biology and bioinformatics. The spatial atomic structures of Fic proteins were determined by X-ray crystallographic analyses carried out by the Schirmer Group at the Swiss Light Source (in Villigen) revealing the detailed geometry of the active center of the enzyme along with the inhibiting glutamate-finger. Dehio’s group, on the other hand, could demonstrate the inhibitory role of this glutamate-finger by using a combination of functional studies and mutagenesis and showed the overall significance of the discovery through extensive protein sequence comparisons.
Based on the gained insights, it has now become possible to investigate the biological role of many members of the vast Fic family. Furthermore, with this knowledge, scientists will now be able to search for the physiological signal of Fic activation in a directed way.
Original article
Philipp Engel, Arnaud Goepfert, Frédéric V. Stanger, Alexander Harms, Alexander Schmidt, Tilman Schirmer & Christoph Dehio
Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins
Nature, published online 22 January 2012 | doi: 10.1038/nature10729
Further Information
Prof. Dr. Christoph Dehio, Biozentrum, University of Basel, Tel. 061 267 21 40, Email: christoph.dehio@unibas.ch
Prof. Dr. Tilman Schirmer, Biozentrum, University of Basel, Tel. 061 267 28 89, Email: tilman.schirmer@unibas.ch
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Superradiant atoms could push the boundaries of how precisely time can be measured
Superradiant atoms can help us measure time more precisely than ever. In a new study, researchers from the University of Copenhagen present a new method for measuring the time interval,…

Ion thermoelectric conversion devices for near room temperature
The electrode sheet of the thermoelectric device consists of ionic hydrogel, which is sandwiched between the electrodes to form, and the Prussian blue on the electrode undergoes a redox reaction…

Zap Energy achieves 37-million-degree temperatures in a compact device
New publication reports record electron temperatures for a small-scale, sheared-flow-stabilized Z-pinch fusion device. In the nine decades since humans first produced fusion reactions, only a few fusion technologies have demonstrated…





















