Big data approach to predict protein structure
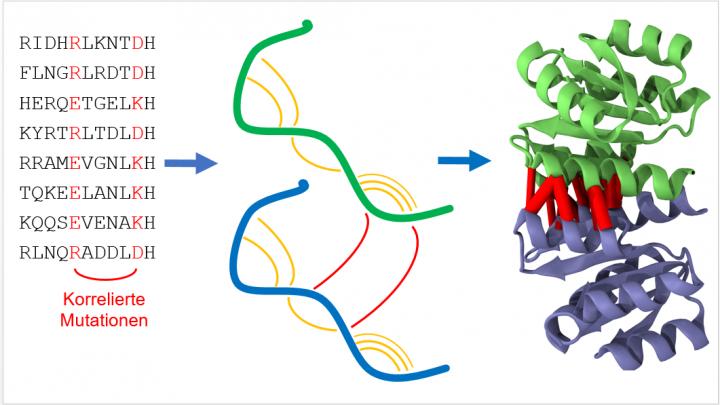
Homodimers are identical pairs of protein chains (proteins, green and blue) that bind to each other. Statistical analysis of protein sequences looks for mutations reflecting spatial proximity of protein segments both within the same protein (orange) and with a partner protein (red). This information can be used to predict the protein structure of the homodimer. Photo: KIT
Nothing works without proteins in the body, they are the molecular all-rounders in our cells. If they do not work properly, severe diseases, such as Alzheimer's, may result. To develop methods to repair malfunctioning proteins, their structure has to be known. Using a big data approach, researchers of Karlsruhe Institute of Technology (KIT) have now developed a method to predict protein structures.
In the Proceedings of the National Academy of Sciences of the United States of America (PNAS), the researchers report that they succeeded in predicting even most complicated protein structures by statistical analyses irrespective of the experiment. Experimental determination of protein structures is quite cumbersome, success is not guaranteed. Proteins are the basis of life. As structural proteins, they are involved in the growth of tissue, such as nails or hairs. Other proteins work as muscles, control metabolism and immune response, or transport oxygen in the red blood cells.
The basic structure of proteins with certain functions is similar in different organisms. “No matter whether human being, mouse, whale or bacterium, nature does not constantly invent proteins for various living organisms anew, but varies them by evolutionary mutation and selection,” Alexander Schug of the Steinbuch Centre for Computing (SCC) says. Such mutations can be identified easily when reading out the genetic information making up the proteins. If mutations occur in pairs, the protein sections involved mostly are located close to each other. With the help of a computer, the data of many spatially adjacent sections can be composed to an exact prediction of the three-dimensional structure similar to a big puzzle. “To understand the function of a protein in detail and to influence it, if possible, the place of every individual atom has to be known,” Schug says.
For his work, the physicist uses an interdisciplinary approach based on methods and resources of computer science and biochemistry. Using supercomputers, he searched the freely available genetic information of thousands of organisms, ranging from bacteria to the human being, for correlated mutations. “By combining latest technology and a true treasure of datasets, we studied nearly two thousand different proteins. This is a completely new dimension compared to previous studies,” Schug adds. He emphasizes that this shows the high performance of the method that promises to be of high potential for applications ranging from molecular biology to medicine. Although present work is fundamental research according to Schug, the results may well be incorporated in new treatment methods of diseases in the future.
###
Karlsruhe Institute of Technology (KIT) pools its three core tasks of research, higher education, and innovation in a mission. With about 9,300 employees and 25,000 students, KIT is one of the big institutions of research and higher education in natural sciences and engineering in Europe.
KIT – The Research University in the Helmholtz Association
Media Contact
All latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

Properties of new materials for microchips
… can now be measured well. Reseachers of Delft University of Technology demonstrated measuring performance properties of ultrathin silicon membranes. Making ever smaller and more powerful chips requires new ultrathin…

Floating solar’s potential
… to support sustainable development by addressing climate, water, and energy goals holistically. A new study published this week in Nature Energy raises the potential for floating solar photovoltaics (FPV)…

Skyrmions move at record speeds
… a step towards the computing of the future. An international research team led by scientists from the CNRS1 has discovered that the magnetic nanobubbles2 known as skyrmions can be…





















