Adding to the list of disease-causing proteins in brain disorders
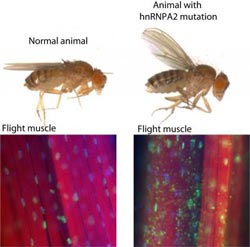
This shows the wing posture in a normal fly and a fly with hnRNPA2 mutation. The mutant fly is weak and unable to maintain normal wing posture. Muscle histology shows that hnRNPA2 is nuclear in normal flies, but accumulates in innumerable cytoplasmic aggregates in flies with the hnRNPA2 mutation.<br><br>Credit: Nam Chul Kim, St. Jude Children’s Research Hospital; Nature<br>
A multi-institution group of researchers has found new candidate disease proteins for neurodegenerative disorders.
James Shorter, Ph.D., assistant professor of Biochemistry and Biophysics at the Perelman School of Medicine, University of Pennsylvania, Paul Taylor, M.D., PhD, St. Jude Children's Research Hospital, and colleagues describe in an advanced online publication of Nature that mutations in prion-like segments of two RNA-binding proteins are associated with a rare inherited degeneration disorder affecting muscle, brain, motor neurons and bone (called multisystem proteinopathy) and one case of the familial form of amyotrophic lateral sclerosis (ALS).
“This study uses a variety of scientific approaches to provide powerful evidence that unregulated polymerization of proteins involved in RNA metabolism may contribute to ALS and related diseases,” said Amelie Gubitz, Ph.D., a program director at the National Institute of Neurological Disorders and Stroke (NINDS).
ALS, or Lou Gehrig's disease, is a universally fatal neurodegenerative disease. Previous studies found that mutations in two related RNA-binding proteins, TDP-43 and FUS, cause some forms of ALS, but more proteins were suspected of causing other forms of the disease. TDP-43 and FUS regulate how the genetic code is translated for the assembly of proteins.
There are over 200 human RNA-binding proteins, including FUS and TDP-43, raising the possibility that additional RNA-binding proteins might contribute to ALS pathology. Computer algorithms, based on protein sequences, designed to identify yeast prions predict that around 250 human proteins, including several RNA-binding proteins associated with neurodegenerative disease, harbor a distinctive prion-like segment. These segments are essential for the assembly of certain protein complexes. But, the interplay between human prion-like segments and disease is not well understood.
Using yeast as a model organism, co-author Aaron Gitler, while at Penn in 2011, surveyed 133 of 200-plus candidate human RNA-binding proteins to predict new ALS disease genes, other than TDP-43 and FUS. They further winnowed the candidates to about 10 proteins with prion-like segments, and selected two candidates, TAF15 and EWSR1, for further study. Both TAF15 and EWSR1 aggregated in the test tube and were toxic in yeast.
Remarkably, they also uncovered TAF15 and EWSR1 mutations in ALS patients that were not found in healthy individuals. Based on these findings, they proposed that RNA-binding proteins with prion-like segments might contribute very broadly to the pathology of ALS and related brain disorders.
Characterizing the Top-Ten
Taylor, Gitler, Shorter, and others continued to characterize the top-ten human RNA-binding proteins with prion-like segments. The Nature study describes that two more of the top-ten candidates, called hnRNPA1 and hnRNPA2B1, are mutated and cause familial cases of brain disease. The mutations in hnRNPA1 and hnRNPA2B1 were present in two families with an extremely rare inherited degeneration affecting muscle, brain, motor neuron, and bone and another from a person with familial ALS.
Mutations in these two proteins fell in the prion-like segments and coincided with “sticky” regions in the proteins, making these regions more prone to assemble into self-organizing fibrils. The normal form of the proteins shows a natural tendency to assemble into fibrils, which is exacerbated by the disease mutations.
“The mutations accelerate the formation of the fibrils that recruit normal protein to form more fibrils,” noted co-first author Emily Scarborough, from Penn. This dysregulated assembly likely contributes to disease. Indeed, the disease mutations also promote excess incorporation of the proteins into stress granules within a cell and the formation of clumps in the cells of animal models of human neurodegenerative disease.
“Neurodegenerative disease could ensue from unregulated fibril formation initiated spontaneously by environmental stress or another factor that regulates a protein's assembly,” says Scarborough.
“This paper reflects an amazing collaborative effort and provides a great example of how understanding the underlying pure protein biochemistry can help explain how genetic mutations might cause pathology and disease,” says Shorter.
“The findings confirm a strong prediction that the disease-causing mutations make the prion-like segment 'stickier' and more prone to clump,” added co-first author Zamia Diaz, also from Penn.
Diseases associated with fibrils forming from prion-like domains in proteins frequently show “spreading” pathology, in which cellular degeneration via inclusions starts in one center of the brain and “spreads” to neighboring tissue. Although not directly addressed in the Nature study, the findings suggest that cell-to-cell transmission of a self-templating protein could contribute to the spreading pathology that is characteristic of these diseases.
“Related proteins with prion-like domains must be considered candidates for initiating and perhaps propagating similar pathologies in muscle, brain, motor neurons, and bone,” concluded Shorter.
The research was funded in part by the by Ellison Medical Foundation, an NIH Innovator Fund award, NINDS (DP2OD002177, NS067354), and The Packard Center for ALS Research at Johns Hopkins.
Penn Medicine is one of the world's leading academic medical centers, dedicated to the related missions of medical education, biomedical research, and excellence in patient care. Penn Medicine consists of the Raymond and Ruth Perelman School of Medicine at the University of Pennsylvania (founded in 1765 as the nation's first medical school) and the University of Pennsylvania Health System, which together form a $4.3 billion enterprise.
The Perelman School of Medicine is currently ranked #2 in U.S. News & World Report's survey of research-oriented medical schools. The School is consistently among the nation's top recipients of funding from the National Institutes of Health, with $479.3 million awarded in the 2011 fiscal year.
The University of Pennsylvania Health System's patient care facilities include: The Hospital of the University of Pennsylvania — recognized as one of the nation's top “Honor Roll” hospitals by U.S. News & World Report; Penn Presbyterian Medical Center; and Pennsylvania Hospital — the nation's first hospital, founded in 1751. Penn Medicine also includes additional patient care facilities and services throughout the Philadelphia region.
Penn Medicine is committed to improving lives and health through a variety of community-based programs and activities. In fiscal year 2011, Penn Medicine provided $854 million to benefit our community.
Media Contact
More Information:
http://www.uphs.upenn.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles

“Nanostitches” enable lighter and tougher composite materials
In research that may lead to next-generation airplanes and spacecraft, MIT engineers used carbon nanotubes to prevent cracking in multilayered composites. To save on fuel and reduce aircraft emissions, engineers…

Trash to treasure
Researchers turn metal waste into catalyst for hydrogen. Scientists have found a way to transform metal waste into a highly efficient catalyst to make hydrogen from water, a discovery that…
Real-time detection of infectious disease viruses
… by searching for molecular fingerprinting. A research team consisting of Professor Kyoung-Duck Park and Taeyoung Moon and Huitae Joo, PhD candidates, from the Department of Physics at Pohang University…





















