Just a Little Squeeze Lets Proteins Assess DNA

Scientists had thought DNA-binding proteins primarily used full-body hugs for accurate readings of the information coded in the DNA's sequence.
Even a protein known to use the hug method, called direct readout, can effectively pinpoint sites on DNA using indirect readout, found researcher Nancy C. Horton and her colleagues.
“It was a total surprise,” said Horton, a UA associate professor of biochemistry and molecular biophysics. “No one had ever seen it before.”
Doing the quick squeezes that scientists call indirect readout probably works faster than requiring full-body contact with all the DNA, the researchers suggest. Quick and accurate identification of key sites on DNA is important for the health of all kinds of cells, from bacteria to humans.
To detect the protein-DNA connection in such detail, Horton and her co-authors Elizabeth J. Little and Andrea C. Babic studied a DNA-binding protein that bacteria use to protect themselves from viral infections.
The finding has implications for the development of designer drugs.
“People have and are developing DNA-binding proteins to turn genes on and off,” Horton said. Such designer proteins can be used to cut out the bad copy of a gene and help replace it with good copy.
“We found that indirect readout is important for finding the right sequence, and we now think indirect readout is also important for finding it quickly,”
she said.
The team published their paper, “Early Interrogation and Recognition of DNA Sequence by Indirect Readout,” in the December issue of the journal Structure. First author Little and co-author Babic were postdoctoral research associates in Horton's laboratory when they did the research. The two are now senior scientists at Ventana Medical Systems, Inc. in Tucson, Ariz.
The National Institutes of Health funded the research.
Horton studies proteins that bind to DNA.
Seven years ago, she figured out the structure of a protein called HincII that snips up DNA. The protein is a type of enzyme called a restriction endonuclease and comes from Haemophilus influenzae bacteria.
Since that time, Horton has been trying to learn how HincII interrogates the DNA to find the right place to cut.
The protein protects bacteria by cutting up DNA from invading viruses.
Without the protective protein, viral DNA would commandeer the bacterium's cellular machinery to produce viruses and ultimately kill the bacterial cell.
The HincII protein distinguishes between bacterial DNA and viral DNA by recognizing certain sequences on DNA. Such a defense requires speed to prevent the marauding virus from killing the cell and also accuracy so the protein doesn't accidentally hack up the bacterium's own DNA.
Horton knew from her previous work that the HincII protein used the direct readout method to find the particular sequence of DNA that corresponded to enemy DNA. The protein seemed to distort the DNA to read it.
Removing the direct readout contact between the protein and the DNA might show whether the DNA distortion or the contact itself was important, Horton said.
Therefore Little and Babic created a mutant protein that couldn't hug DNA closely and therefore couldn't use the direct readout method. Little described the mutant protein as missing the fingers the normal protein used to probe the DNA.
“If the finger was doing all the recognition, then the mutant should cut any DNA sequence,” Horton said.
To see how the mutant interacted with DNA, the researchers crystallized the mutant protein-DNA complex in action.
Initially, Horton thought the assay had gone wrong and almost threw the results in the trash, she wrote in an e-mail.
The mutant protein had chosen the proper site on the DNA with 100 percent specificity, which was opposite from what she expected. In addition, the DNA was distorted, even though the mutant couldn't make the strong contact a normal protein would.
“I did a double-take. I was just taking a picture to have a record that it was non-specific,” she said.
Understanding how endonucleases and other DNA binding-proteins recognize a particular DNA sequence provides insight into key cellular processes including the replication, transcription and repair of DNA.
Little said, “In every single one of your cells are proteins looking for the proper sequences in DNA in order to make the proteins you need to stay alive.”
Horton added, “Understanding how these processes work helps in the understanding of diseases so that we could potentially cure the disease.”
Researcher contact information:
Nancy C. Horton
Associate Professor of Biochemistry and Molecular Biophysics nhorton@u.arizona.edu
520-626-3828 (office)
520-626-0246 (lab)
Media Contact
More Information:
http://www.arizona.eduAll latest news from the category: Life Sciences and Chemistry
Articles and reports from the Life Sciences and chemistry area deal with applied and basic research into modern biology, chemistry and human medicine.
Valuable information can be found on a range of life sciences fields including bacteriology, biochemistry, bionics, bioinformatics, biophysics, biotechnology, genetics, geobotany, human biology, marine biology, microbiology, molecular biology, cellular biology, zoology, bioinorganic chemistry, microchemistry and environmental chemistry.
Newest articles
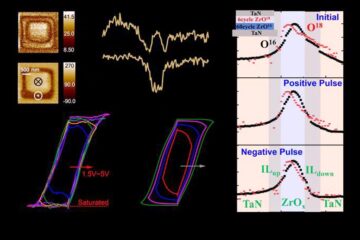
Evidence for reversible oxygen ion movement during electrical pulsing
…enabler of the emerging ferroelectricity in binary oxides. In a recent study published in Materials Futures, researchers have uncovered a pivotal mechanism driving the emergence of ferroelectricity in binary oxides….
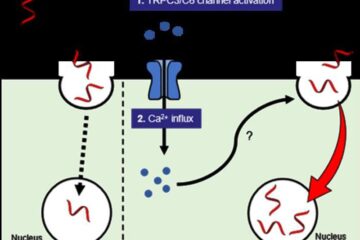
Next-generation treatments hitch a ride into cancer cells
Researchers from Osaka University discover that opening a channel into cancer cells helps antisense oligonucleotide drugs reach their targets. Antisense oligonucleotides (ASOs) are next-generation drugs that can treat disease by…

Boron deficiency: oilseed rape reacts as with infection and pest infestation
Genetic mechanisms uncovered… Boron deficiency has a devastating effect on oilseed rape and related plants. However, little is known about the underlying genetic mechanisms. A study shows that the response…





















