DNA blueprint for healthier and more efficient cows

The findings, published in two reports in the journal Science today, will have a profound impact on Australia’s livestock industry.
CSIRO scientists were among the 300 researchers from 25 countries involved in the six-year Bovine Genome Sequencing Project designed to sequence, annotate and analyse the genome of a female Hereford cow called L1 Dominette.
The scientists discovered that the bovine genome contains 2,870 billion DNA building blocks, encoding a minimum of 22,000 genes. Of major interest to scientists are the differences in the organisation of the genes involved in lactation, reproduction, digestion and metabolism in cows compared to other mammals.
One of the lead authors of the report on the project’s latest findings, CSIRO Livestock Industries researcher Dr Ross Tellam said the bovine genome has about 14,000 genes which are common to all mammals and these constitute the ‘engine room’ of mammalian biology.
“The team found that cows share about 80 per cent of their genes with humans, also providing us with a better understanding of the human genome,” Dr Tellam said.
“One of the surprises in the analysis was that cow and human proteins have more in common than mouse and human proteins, yet it is the mouse that is often used in medical research as a model of human disease conditions.”
Dr Tellam said the research provides an insight into the unique biology and evolution of ruminant animals and helps explain why they have been so successful as a species.
One of the major findings was that the cow has significant rearrangements in many of its immune genes and presumably an enhanced natural ability to defend itself from disease.
“This may be an evolutionary response to an increased risk of opportunistic infections at mucosal surfaces caused by the large number of bacteria and fungi carried in the rumen (the largest of the four compartments that make up the bovine stomach),” Dr Tellam said.
“The second possible explanation is that ruminants and cows are typically found in very large herds, and in these herds there is a greater propensity for disease transmission, so you need to be better equipped to withstand diseases.”
These new findings will point the way for future research that could result in more sustainable food production.
Dr Tellam said the $US53 million Bovine Genome Sequencing Project – led by the Human Genome Sequencing Centre at Baylor College of Medicine (BCM-HGSC) in Houston, Texas – is an example of major achievements that can only be realised by substantial international scientific cooperation.
Using the complete genome sequence from L1 Dominette, the female Hereford cow, scientists also undertook comparative genome sequencing for six more breeds to look for genetic changes.
The resulting bovine HapMap – a literal map of genetic diversity among different populations – is also published in today’s edition of the journal Science.
“Domestication and artificial selection appear to have left detectable signatures of selection within the cattle genome yet the current level of diversity within breeds is at least as great as that found within humans,” CSIRO Livestock Industries scientist and one of the project’s group leaders, Dr Bill Barendse, said.
The implications of the genome project for the beef and dairy industries are enormous.
“The availability of very large numbers of single nucleotide polymorphisms (DNA changes in the genetic blueprint) has allowed the development of gene chips that measure genetic variation in cattle populations and will allow the rapid selective breeding of animals with higher value commercial traits.
“This technology is quickly transforming the dairy genetics industry and has the potential to dramatically alter beef cattle industries as well,” Dr Barendse said.
These new genetic tools may provide a means to select more energy-efficient animals with a smaller environmental footprint, particularly animals that produce less greenhouse gas.
The Bovine Sequencing and Analysis Project was led by Drs Richard Gibbs and George Weinstock, co-directors of the BCM-HGSC, Dr Steven Kappes of the US Department of Agriculture, Dr Christine Elsik of Georgetown University and Dr Ross Tellam of CSIRO Australia.
In addition to CSIRO, major funders of the Project were: the National Human Genome Research Institute, which funded more than half of the project; the U.S. Department of Agriculture's Agricultural Research Service and Cooperative State Research, Education, and Extension Service National Research Initiative; the state of Texas; Genome Canada through Genome British Columbia; The Alberta Science and Research Authority; Agritech Investments Ltd., Dairy Insight, Inc. and AgResearch Ltd., all of New Zealand; the Research Council of Norway; the Kleberg Foundation; and the National, Texas and South Dakota Beef Check-off Funds.
The Bovine HapMap Project was led by Drs Richard Gibbs and Curt Van Tassell of the USDA and Dr Jeremy Taylor of the University of Missouri.
Funding for the Bovine HapMap Project was provided by: American Angus Association, American Hereford Association, American Jersey Cattle Association, AgResearch (New Zealand), Beef CRC and Meat and Livestock Australia for the Australian Brahman Breeders Association, Beefmaster Breeders United, The Brazilian Agricultural Research Corporation (Embrapa), Brown Swiss Association, GENO Breeding and Artificial Insemination Association – Norway, Herd Book/France Limousin Selection, Holstein Association USA, International Atomic Energy Agency – FAO/IAEA Vienna, International Livestock Research Institute – Kenya, Italian Piedmontese Breeders – Parco Tecnologico Padano, Italian Romagnola Society – Università Cattolica del Sacro Cuore, Livestock Improvement Corporation, Meat & Wool New Zealand. North American Limousin Foundation, Red Angus Association of America, The Roslin Institute for UK Guernsey, and Sygen (now Genus).
Media Contact
More Information:
http://www.csiro.auAll latest news from the category: Agricultural and Forestry Science
Newest articles
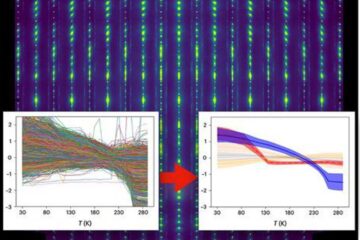
Machine learning algorithm reveals long-theorized glass phase in crystal
Scientists have found evidence of an elusive, glassy phase of matter that emerges when a crystal’s perfect internal pattern is disrupted. X-ray technology and machine learning converge to shed light…
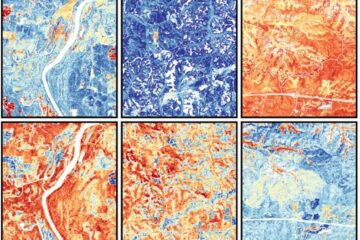
Mapping plant functional diversity from space
HKU ecologists revolutionize ecosystem monitoring with novel field-satellite integration. An international team of researchers, led by Professor Jin WU from the School of Biological Sciences at The University of Hong…

Inverters with constant full load capability
…enable an increase in the performance of electric drives. Overheating components significantly limit the performance of drivetrains in electric vehicles. Inverters in particular are subject to a high thermal load,…





















